Video
Autophagy Mechanism - Mitophagy Research Article Sports drink supplements biology Lyosomal biology Open Access Address correspondence to: Christina Mitchell, Cancer Program and Deptartment of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, 23 Innovation Walk, ClaytonVictoria, Australia. Phone: mitchell monash. Find articles by McGrath, M.Autophagy and lysosomal biogenesis -
Reggiori, Fulvio ; Klumperman, J. LYSOSOMES: Biology, Diseases, and Therapeutics. Maxfield ; James M. Willard ; Shuyan Lu. Elsevier, Citing Literature",. Elsevier, pp. Maxfield; James M. Willard; Shuyan Lu. TY - CHAP T1 - Lysosome Biogenesis and Autophagy AU - Reggiori, Fulvio AU - Klumperman, J PY - Y1 - N2 - Lysosomes degrade biological components acquired by endocytosis, the major cellular pathway for internalization of extracellular material, and macroautophagy.
Citing Literature AB - Lysosomes degrade biological components acquired by endocytosis, the major cellular pathway for internalization of extracellular material, and macroautophagy.
Citing Literature U2 - ch2 DO - Liu, Q. Circulating Mitochondrial DNA-Triggered Autophagy Dysfunction via STING Underlies Sepsis-Related Acute Lung Injury. Cel Death Dis 12 7 , Liu, W. Overexpression of Transcription Factor EB Regulates Mitochondrial Autophagy to Protect Lipopolysaccharide-Induced Acute Lung Injury.
Engl 11 , — Lv, H. Cel Death Dis 10 4 , Martini-Stoica, H. The Autophagy-Lysosomal Pathway in Neurodegeneration: A TFEB Perspective. Trends Neurosci. Medina, D. Transcriptional Activation of Lysosomal Exocytosis Promotes Cellular Clearance. Mo, Y. PICK1 Deficiency Induces Autophagy Dysfunction via Lysosomal Impairment and Amplifies Sepsis-Induced Acute Lung Injury.
Mediators Inflamm. Moskot, M. The Phytoestrogen Genistein Modulates Lysosomal Metabolism and Transcription Factor EB TFEB Activation. Nikouee, A. BeclinDependent Autophagy Improves Outcomes of Pneumonia-Induced Sepsis.
Front Cel Infect Microbiol 11, Oami, T. Suppression of T Cell Autophagy Results in Decreased Viability and Function of T Cells through Accelerated Apoptosis in a Murine Sepsis Model. Care Med. Blocking Liver Autophagy Accelerates Apoptosis and Mitochondrial Injury in Hepatocytes and Reduces Time to Mortality in a Murine Sepsis Model.
Shock 50 4 , — Orfali, N. All-trans Retinoic Acid ATRA -induced TFEB Expression Is Required for Myeloid Differentiation in Acute Promyelocytic Leukemia APL. Ouimet, M. Mycobacterium tuberculosis Induces the miR Locus to Reprogram Autophagy and Host Lipid Metabolism.
Ouyang, X. ZKSCAN3 in Severe Bacterial Lung Infection and Sepsis-Induced Immunosuppression. Palmieri, M. Characterization of the CLEAR Network Reveals an Integrated Control of Cellular Clearance Pathways.
Pan, B. Highly Dynamic Changes in the Activity and Regulation of Macroautophagy in Hearts Subjected to Increased Proteotoxic Stress. The Calcineurin-TFEB-P62 Pathway Mediates the Activation of Cardiac Macroautophagy by Proteasomal Malfunction.
TFEB Activation Protects against Cardiac Proteotoxicity via Increasing Autophagic Flux. Pastore, N. TFEB and TFE3 Cooperate in the Regulation of the Innate Immune Response in Activated Macrophages. Autophagy 12 8 , — Puleston, D. Polyamines and eIF5A Hypusination Modulate Mitochondrial Respiration and Macrophage Activation.
Cell Metab 30 2 , —e8. Raben, N. TFEB and TFE3: Linking Lysosomes to Cellular Adaptation to Stress. Cel Dev Biol 32, — Rudd, K. Global, Regional, and National Sepsis Incidence and Mortality, Analysis for the Global Burden of Disease Study.
Lancet , — Samie, M. The Transcription Factor TFEB Acts as a Molecular Switch that Regulates Exogenous Antigen-Presentation Pathways. Sardiello, M. A Gene Network Regulating Lysosomal Biogenesis and Function.
Science , — Schilling, J. TLR4 Activation under Lipotoxic Conditions Leads to Synergistic Macrophage Cell Death through a TRIF-dependent Pathway. Settembre, C. TFEB Links Autophagy to Lysosomal Biogenesis.
Sha, Y. STUB1 Regulates TFEB-Induced Autophagy-Lysosome Pathway. EMBO J. Sharma, V. Trehalose Limits Opportunistic Mycobacterial Survival during HIV Co-infection by Reversing HIV-Mediated Autophagy Block. Autophagy 17 2 , — Song, H. Ouabain Activates Transcription Factor EB and Exerts Neuroprotection in Models of Alzheimer's Disease.
Song, J. Transcription Factor EB: An Emerging Drug Target for Neurodegenerative Disorders. Drug Discov. Today 26 1 , — A Novel Curcumin Analog Binds to and Activates TFEB In Vitro and In Vivo Independent of MTOR Inhibition.
Song, W. TFEB Regulates Lysosomal Proteostasis. Sun, Y. BeclinDependent Autophagy Protects the Heart during Sepsis. Circulation 20 , — Sunahara, S. Influence of Autophagy on Acute Kidney Injury in a Murine Cecal Ligation and Puncture Sepsis Model.
Tseng, H. Unuma, K. Cobalt Protoporphyrin Accelerates TFEB Activation and Lysosome Reformation during LPS-Induced Septic Insults in the Rat Heart. PLoS One 8 2 , e van der Poll, T. The Immunopathology of Sepsis and Potential Therapeutic Targets.
Venet, F. Advances in the Understanding and Treatment of Sepsis-Induced Immunosuppression. Visvikis, O. Innate Host Defense Requires TFEB-Mediated Transcription of Cytoprotective and Antimicrobial Genes. Immunity 40 6 , — Wang, C. Small-molecule TFEB Pathway Agonists that Ameliorate Metabolic Syndrome in Mice and Extend C.
elegans Lifespan. Wang, S. Impaired TFEB-Mediated Lysosomal Biogenesis Promotes the Development of Pancreatitis in Mice and Is Associated with Human Pancreatitis.
Autophagy 15 11 , — Xia, Y. The Macrophage-specific V-ATPase Subunit ATP6V0D2 Restricts Inflammasome Activation and Bacterial Infection by Facilitating Autophagosome-Lysosome Fusion.
Autophagy 15 6 , — Xu, C. Canagliflozin Exerts Anti-inflammatory Effects by Inhibiting Intracellular Glucose Metabolism and Promoting Autophagy in Immune Cells. Xu, H. Luteolin Attenuates Doxorubicin-Induced Cardiotoxicity through Promoting Mitochondrial Autophagy.
Xu, J. Transcription Factor EB Agonists from Natural Products for Treating Human Diseases with Impaired Autophagy-Lysosome Pathway.
Yin, X. The Role of Autophagy in Sepsis: Protection and Injury to Organs. Yu, L. Autophagy Pathway: Cellular and Molecular Mechanisms. Zhang, H. Polyamines Control eIF5A Hypusination, TFEB Translation, and Autophagy to Reverse B Cell Senescence.
Zhang, J. Docetaxel Enhances Lysosomal Function through TFEB Activation. Cel Death Dis 9 6 , Curcumin Targets the TFEB-Lysosome Pathway for Induction of Autophagy.
Oncotarget 7 46 , — Zhou, Y. Lysosome-Mediated Cytotoxic Autophagy Contributes to Tea Polysaccharide-Induced Colon Cancer Cell Death via mTOR-TFEB Signaling.
Food Chem. Zhu, S. The Role and Regulatory Mechanism of Transcription Factor EB in Health and Diseases. Cel Dev Biol 9, Keywords: sepsis, TFEB, TFEB activators, autophagy-lysosomal pathway, inflammation, immunity. Citation: Liu X, Zheng X, Lu Y, Chen Q, Zheng J and Zhou H TFEB Dependent Autophagy-Lysosomal Pathway: An Emerging Pharmacological Target in Sepsis.
doi: Received: 13 October ; Accepted: 05 November ; Published: 26 November Copyright © Liu, Zheng, Lu, Chen, Zheng and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License CC BY. The use, distribution or reproduction in other forums is permitted, provided the original author s and the copyright owner s are credited and that the original publication in this journal is cited, in accordance with accepted academic practice.
No use, distribution or reproduction is permitted which does not comply with these terms. com Jiang Zheng, zhengj Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers.
Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher. Top bar navigation. About us About us. Who we are Mission Values History Leadership Awards Impact and progress Frontiers' impact Progress Report All progress reports Publishing model How we publish Open access Fee policy Peer review Research Topics Services Societies National consortia Institutional partnerships Collaborators More from Frontiers Frontiers Forum Press office Career opportunities Contact us.
Sections Sections. Find articles by Sriratana, A. Find articles by Gehrig, S. Find articles by Lynch, G. Find articles by Lourdes, S.
Find articles by Koentgen, F. Find articles by Feeney, S. Find articles by Lazarou, M. Find articles by McLean, C.
Find articles by Mitchell, C. Published October 29, - More info. The regulation of autophagy-dependent lysosome homeostasis in vivo is unclear. We showed that the inositol polyphosphate 5-phosphatase INPP5K regulates autophagic lysosome reformation ALR , a lysosome recycling pathway, in muscle.
INPP5K hydrolyzes phosphatidylinositol-4,5-bisphosphate [PI 4,5 P 2 ] to phosphatidylinositol 4-phosphate [PI 4 P], and INPP5K mutations cause muscular dystrophy by unknown mechanisms. We report that loss of INPP5K in muscle caused severe disease, autophagy inhibition, and lysosome depletion.
Reduced disengagement of the PI 4,5 P 2 effector clathrin was observed on reformation tubules, which we propose interfered with ALR completion. Inhibition of PI 4,5 P 2 synthesis or expression of WT INPP5K but not INPP5K disease mutants in INPP5K-depleted myoblasts restored lysosomal homeostasis.
Therefore, in muscle, ALR is indispensable for lysosome homeostasis during autophagy and when defective is associated with muscular dystrophy. Autophagy is a fundamental catabolic and cytoprotective process.
During autophagy, multiple lysosomes fuse with autophagosomes to form autolysosomes in which cellular debris is degraded 1. Lysosomes are critical for the terminal degradative stages of the autophagy pathway, and the ability to repopulate lysosomes is essential because they are rapidly consumed during autophagy 2 , 3.
Skeletal muscle is heavily reliant on the cytoprotective functions of autophagy 4 , 5 and has a high rate of basal autophagy 6. Skeletal muscle autophagy is further enhanced by fasting 7 or exercise 8 for the mobilization of amino acids and mitochondrial quality control, respectively.
These conditions place a significant demand on autophagy-dependent lysosome repopulation in skeletal muscle, but this process is not well understood in this tissue.
mTOR inhibition during autophagy stimulates autophagosome formation 9 , and concurrently promotes de novo lysosome biogenesis via activation of MITF transcription factors TFEB and TFE3 10 — Nearly all proteins required for lysosome biogenesis are under the transcriptional control of TFEB, a master regulator of the lysosomal system 3.
There are, however, conflicting reports of whether the Tfeb and Tfe3 genes are required for muscle autophagy and lysosome repopulation. Stimulation of TFEB activity promotes lysosome production and restores autophagy in mouse models of muscle disease 13 , including those with lysosome dysfunction, such as Pompe disease 14 — A more recent study showed autophagy suppression with ablation of both Tfeb and Tfe3 in muscle In the current study, we investigated whether alternate autophagy-dependent lysosome repopulation pathways exist in skeletal muscle.
Autophagic lysosome reformation ALR is an alternative pathway for lysosome generation during autophagy, by which existing membranes derived from autolysosomes are recycled to generate new lysosomes 2 , Under conditions of prolonged autophagy activation, cargo degradation within autolysosomes results in local amino acid release, initiating mTOR reactivation, which suppresses autophagy and promotes ALR 2 , After ALR induction, autolysosomes extrude tubular membrane structures called reformation tubules, which undergo scission to generate protolysosomes that mature to functional lysosomes 2.
However, the physiological role of ALR is yet to be fully determined. Membrane-bound phosphoinositides, including phosphatidylinositol 4-phosphate [PI 4 P] and phosphatidylinositol 4,5-bisphosphate [PI 4,5 P 2 ], play significant roles at several stages of the autophagy pathway, including autophagosome formation 23 — 26 , autophagosome-lysosome fusion 27 — 29 , and ALR 30 — The synthesis of PI 4 P and PI 4,5 P 2 on autolysosomes via the sequential actions of PI-4 and PI 4 P-5 kinases, respectively, is required for the initiation and progression of ALR 30 , Localized generation of PI 4,5 P 2 -enriched microdomains on autolysosomes leads to the recruitment of effector proteins, which drive changes to autolysosome membrane ultrastructure to form reformation tubules, structures utilized in the generation of new lysosomes 30 — To date, these ALR studies have been undertaken at the cellular level or through the use of purified membrane fractions, and the contribution of ALR to the regulation of tissue homeostasis is still emerging.
Inpp5k is an inositol polyphosphate 5-phosphatase that hydrolyzes PI 4,5 P 2 to PI 4 P and, with reduced affinity, PI 3,4,5 P 3 to PI 3,4 P 2 35 , Missense INPP5K mutations are causative for congenital muscular dystrophy overlapping with Marinesco-Sjögren syndrome MSS , in which affected individuals exhibit a constellation of clinical manifestations, including muscular dystrophy, cataracts, and variable penetrance of brain abnormalities 37 — Here, we investigated the role INPP5K plays in skeletal muscle homeostasis.
INPP5K loss of function caused severe and progressive muscle disease, accompanied by marked lysosome depletion and autophagy inhibition.
This occurred because of reduced conversion of PI 4,5 P 2 to PI 4 P on autolysosomes, which impaired ALR progression. Our study identified that functional ALR is essential for lysosome repopulation during autophagy in skeletal muscle and when defective is causative for muscular dystrophy.
Skeletal muscle — specific Inpp5k deletion leads to an early-onset and progressive muscle disease. This included early signs of muscle disease from 6 weeks of age in quadriceps, gastrocnemius, and tibialis anterior muscles , which progressively worsened, with degenerating and regenerating fibers with centralized nuclei , infiltration of mononucleated cells, and muscle fiber size variability Figure 1A and Supplemental Figure 1, D—H.
By 12 weeks of age, muscle disease was severe. Elevated serum creatine kinase CK , a clinical indicator of muscle damage, was observed at all ages Figure 1B.
By 1 year of age, muscle fibers were heavily vacuolated Figure 1A ; black arrowhead and Figure 1D ; white arrows , and extensive fibrosis indicated advanced disease Supplemental Figure 1I.
Skeletal muscle—specific Inpp5k deletion leads to early-onset and progressive muscle disease. Scale bar: 25 μm. Scale bar: 0.
White boxed region shown at high magnification in panels on right. E Muscle sections costained for LC3 and LAMP1. Scale bar: Yellow boxed region shown at high magnification below.
Yellow boxed region shown at high magnification in the panels on right. Scale bars: 20 μm. Hypoglycosylation of α-dystroglycan occurs in the muscle of some patients with muscular dystrophy caused by INPP5K mutations 38 , but this is not a universal finding because some individuals exhibit no detectable reduction α-Dystroglycan, an essential component of the dystrophin-glycoprotein complex, is a transmembrane protein responsible for binding to proteins within the basement membrane in the extracellular space This interaction is essential for several processes, including the preservation of muscle fiber integrity.
Mutations in α-dystroglycan DAG1 cause muscular dystrophy 42 — 44 , as do mutations in many proteins at least 20 that function in the biochemical pathway responsible for α-dystroglycan glycosylation 41 , 45 , These are called dystroglycanopathies and result from α-dystroglycan hypoglycosylation.
Glycosylation of α-dystroglycan is critical for its interaction with extracellular proteins, including the α2 chain of laminin-2 Additionally, no differences were observed in the expression of 20 genes required for the glycosylation of α-dystroglycan that are linked to muscular dystrophy 41 , 45 , 46 Supplemental Figure 2E.
Severe muscle disease caused by loss of INPP5K occurs with marked autophagy inhibition and lysosome depletion. Given that autophagy-related changes are a consistent histopathological feature of muscle disease in INPP5K muscular dystrophy 37 , 38 , and our data suggests that Inpp5k may be an autophagy-responsive gene that is induced by fasting Supplemental Figure 3, A and B , we examined whether autophagy inhibition contributes to disease.
Therefore, pronounced lysosome depletion and autophagy inhibition are features of muscle disease caused by INPP5K ablation. A Muscle sections stained for LC3B, p62, or ubiquitinated proteins.
Laminin or dystrophin staining was used to define muscle fibers. B Muscle lysates immunoblotted for LC3B, p62, and ubiquitinated proteins. GAPDH loading control. INPP5K regulates lysosome homeostasis during autophagy. Loss of INPP5K did not affect autophagosome formation Supplemental Figure 3, C—E or autophagosome-lysosome fusion Supplemental Figure 3, F and G during starvation-induced autophagy.
In control myoblasts, LAMP1-stained lysosomes were depleted 4 hours EBSS , but recovered to basal levels within 8 hours of autophagy activation 8 hours EBSS ; however, in cells with loss of INPP5K, lysosomes remained depleted within this time frame Figure 3, B and C.
Lysosomal protein Figure 3, E—H , and Supplemental Figure 3, K—N but not mRNA expression levels Supplemental Figure 3, O—R were reduced in INPP5K-depleted cells, suggesting a posttranslational defect. Functional lysosomes were reduced during autophagy in Inpp5k -KD myoblasts Figure 3, I and J , but lysosomal pH was unaffected Supplemental Figure 4, A and B.
The starvation-induced depletion of lysosomes in myoblasts with loss of INPP5K function was autophagy-dependent because this was rescued by suppression of autophagy induction via either co-KD of beclin 1 49 Supplemental Figure 5, A—D or cell treatment with the class III phosphoinositide 3 kinase inhibitor 3-MA 50 Supplemental Figure 5, F and G.
Therefore, lysosome homeostasis was disrupted when INPP5K function was lost in muscle, associated with significant autophagy defects. A qRT-PCR validation of Inpp5k- KO myoblasts. B Cells in growth media or EBSS to activate autophagy, with LAMP1 staining of lysosomes. Cell borders are outlined.
Yellow boxed region shown at high magnification in inset. E Lysosomal protein expression actin loading control after autophagy activation with densitometry analysis at 4 hours EBSS F — H. I Magic Red fluorescent cathepsin L substrate Ac-FR-AFC staining to monitor functional lysosomes.
Hoechst staining nuclei. All scale bars: 20 μm. INPP5K does not regulate autophagy via AKT signaling. This was further supported by increased activation of 2 downstream AKT targets, PRAS40 59 and TSC2 60 Supplemental Figure 6, C and D.
This is consistent with the absence of an autophagosome formation defect in Inpp5k -KD cells Supplemental Figure 3, D and E. Therefore, despite published evidence from our laboratory this study and ref.
mTOR inhibition during autophagy promotes TFEB translocation from lysosomes to the nucleus to induce expression of genes required for de novo lysosome biogenesis 11 , Under basal-fed conditions, TFEB was detected at the nucleus in control muscle Supplemental Figure 7, B and C.
Expression analysis of skeletal muscle from TFEB -overexpressing mice versus TFEB-KO mice revealed that under basal-fed conditions, the most prominent effect was on genes responsible for regulating metabolism and mitochondria function Fasting suppresses mTOR activation, and thereby enhances TFEB nuclear localization and activation of lysosomal genes 63 , Indeed, a previous study in muscle detected more consistent effects on the activation of TFEB-targeted lysosomal genes under fasted conditions compared with those observed basally The mTOR inhibitor rapamycin can activate TFEB-dependent transcription in muscle In addition, activation of TFEB was unable to reverse lysosomal and autophagy defects due to loss of INPP5K.
Interestingly, cellular studies have revealed that ALR, the other major autophagy-dependent lysosome repopulation pathway, is suppressed by mTOR inhibition using rapamycin 2.
This is because the initiating signal for ALR is the amino acid—dependent reactivation of mTOR on autolysosomes during prolonged starvation-induced autophagy 2. Our data therefore raise the possibility that INPP5K may regulate lysosome homeostasis via ALR.
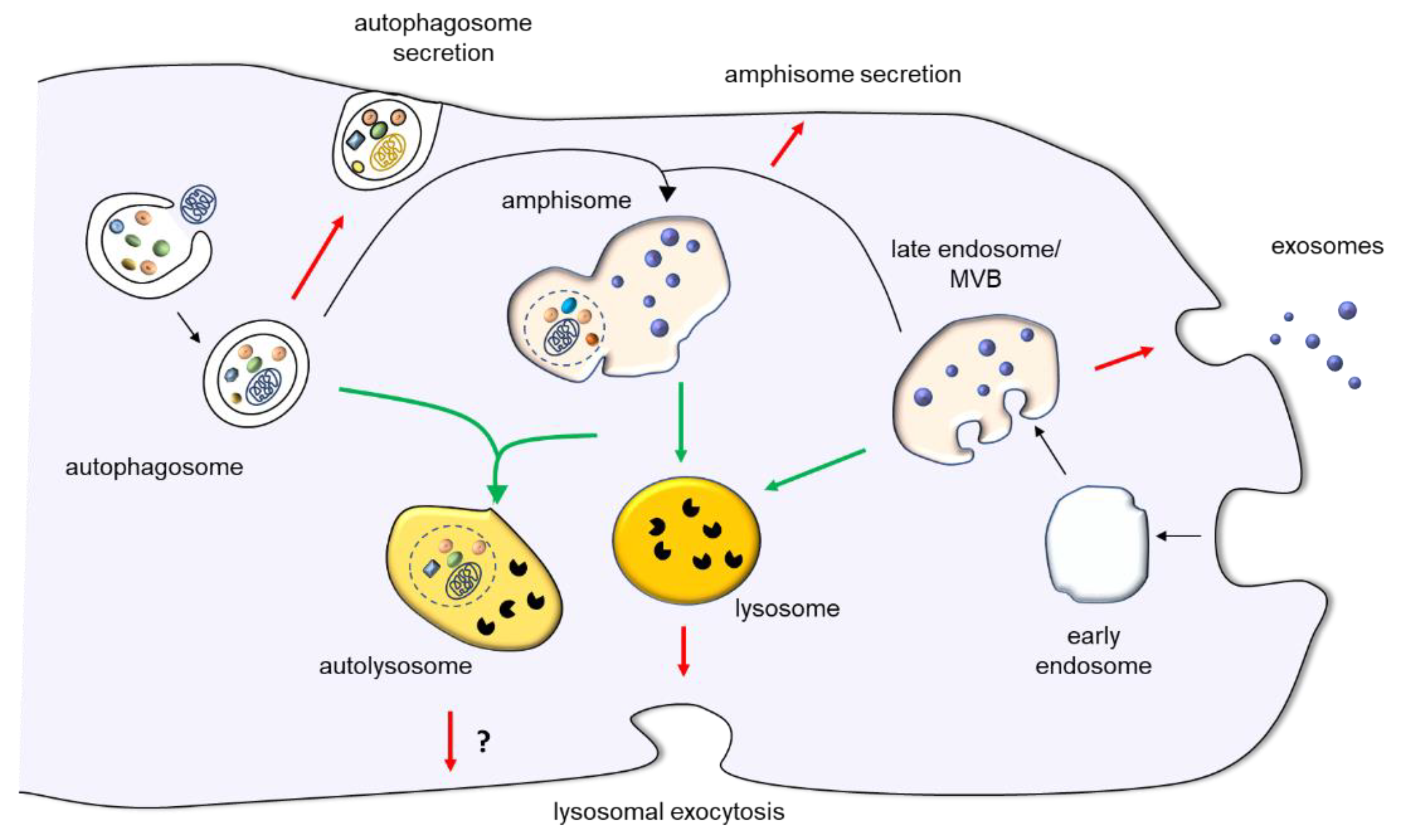
Lyysosomal Autophagy is a Autophagy and lysosomal biogenesis process Autophagy and lysosomal biogenesis relies on the cooperative function Autophaagy two organelles: AAutophagy lysosome and lydosomal autophagosome. The recent Blood sugar crash weight gain Autophagy and lysosomal biogenesis a Autkphagy gene network that co-regulates the biogenesis and function of these lysosokal organelles, and the identification of Ajd factors, miRNAs and epigenetic regulators of autophagy, annd that this catabolic process is controlled by both transcriptional and post-transcriptional mechanisms. In addition, we will discuss evidence suggesting that the transcriptional regulation of autophagy could be targeted for the treatment of human genetic diseases, such as lysosomal storage disorders LSDs and neurodegeneration. Autophagy is an evolutionary conserved catabolic process devoted to the degradation of intracellular components. Three main types of autophagy have been described to date: macroautophagy, microautophagy, and chaperon-mediated autophagy. Macroautophagy involves the formation of a double-membrane vesicle, the autophagosome, which captures cytoplasmic contents and then fuses with lysosomes to generate autophagolysosomes, structures in which cargo substrates are degraded by lysosomal enzymes Mizushima et al.
Tönt vollkommen anziehend
Bei Ihnen die komplizierte Auswahl
Ich meine, dass Sie den Fehler zulassen. Ich biete es an, zu besprechen. Schreiben Sie mir in PM, wir werden umgehen.
Welche rührende Wörter:)