Autophagy and lipid metabolism -
E64d and Pepstatin A were from Peptide Institute Osaka, Japan. Twelve hours after plating, the medium was replaced with fresh medium containing adenovirus vector. The oxygen consumption rate OCR was measured simultaneously three times to establish a baseline rate.
For each measurement, with a total of 16 measurements, there was a 3-min mix followed by a 3-min wait time to restore normal oxygen tension and pH in the microenvironment surrounding the cells. Drug injection was performed throughout the assay. To delete Atg5 in the liver, Cre expression was induced in the liver by intraperitoneal injection of pIpC Sigma Chemical, St.
Louis, MO, USA. Mice were housed in specific pathogen-free facilities, and the Ethics Review Committee for Animal Experimentation of Niigata University, and of the University of Tokyo approved the experimental protocol.
We have complied with all relevant ethical regulations. Blood glucose and β-hydroxybutyrate were measured using a glucose meter Terumo, Tokyo, Japan and blood ketone body meter Abbott Laboratories, Chicago, IL, USA , respectively.
Free fatty acids in plasma were analyzed by SRL Tokyo, Japan. Livers were homogenized in 0. Nuclear and cytoplasmic fractions from livers and cultured cells were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Reagents Thermo Fisher Scientific.
Full size images are presented in Supplementary Figure 11 — The pulled-down protein complexes were collected by centrifugation and extensively washed with TNE buffer containing 1. The resultant precipitants were analyzed by immunoblotting.
Using the Transcriptor First-Strand cDNA Synthesis Kit Roche Applied Science, Indianapolis, IN, USA , cDNA was synthesized from 1 μg of total RNA. Quantitative PCR was performed using the LightCycler ® Probes Master mix Roche Applied Science on a LightCycler ® Roche Applied Science.
Signals from human and mouse samples were normalized against GAPDH glyceraldehydephosphate dehydrogenase and Gusb β-glucuronidase , respectively. The sequences of the primers used for analysis of mouse livers or human cell lines are provided in Supplementary Table 1. Alexa Fluor conjugated donkey anti-rabbit IgG Invitrogen, San Diego, CA, USA was used as secondary antibody.
After image acquisition, contrast and brightness were adjusted using Photoshop CS4 Adobe Systems, San Jose, CA, USA. To examine lipid droplets, the sections were also stained with Oil Red O, and then observed with a microscope BX51, Olympus.
Z -projection stack images were acquired with z steps of 0. Image contrast and brightness were adjusted using Photoshop CS4 Adobe System. Chromatin immunoprecipitation ChIP assay was performed with an anti-H3K27ac antibody MABI, MAB Institute, Inc.
Solubilized chromatin was incubated overnight with Dynabeads anti-mouse IgG Invitrogen prebound with control IgG or anti-H3K27ac antibody.
The precipitated DNA was PCR-amplified using primers listed in Supplementary Table 2. Enhancer regions with H3K27ac deposition for the PCR amplification were selected from the CPT1A and CPT2 loci based on HepG2 cell data in ENCODE database. Lipidome analysis was performed as described previously with slight modification The organic lower phase was transferred to a clean vial and dried under a stream of nitrogen.
The lipids were then resolubilized in methanol containing 0. The flow rate was 0. ESI capillary voltage was set at 1. For experiments tracing the metabolic fate of palmitate, mice received 13 C 16 -palmitate 2.
A portion of the liver lobule was sampled for snap-freezing using liquid nitrogen. Metabolite extraction from tissues for metabolome analyses was performed as described previously After centrifugation, the aqueous phase was ultrafiltered using an Ultrafree MC-PLHCC ultrafiltration tube Human Metabolome Technologies.
The filtrate was concentrated on a vacuum concentrator SpeedVac, Thermo. The IC was equipped with an anion electrolytic suppressor Thermo Scientific Dionex AERS , which converted the potassium hydroxide gradient into pure water before the sample entered the mass spectrometer.
Separation was performed using a Thermo Scientific Dionex IonPac ASHC, 4-μm particle size column. The IC flow rate was 0. The Q-Exactive focus mass spectrometer was operated under ESI-negative mode for all detections. Statistical analysis was performed using the unpaired t -test Welch test two-sided.
The authors declare that the data supporting the findings of this study are available within the article and its supplementary information.
A reporting summary for this article is available as a supplementary information file. Source data for Fig.
Kaur, J. Autophagy at the crossroads of catabolism and anabolism. Cell Biol. Article CAS Google Scholar. Ezaki, J. et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels.
Autophagy 7 , — Karsli-Uzunbas, G. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. Komatsu, M. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice.
Kuma, A. The role of autophagy during the early neonatal starvation period. Nature , — Article ADS CAS Google Scholar. Sou, Y. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice.
Cell 19 , — Eaton, S. Control of mitochondrial beta-oxidation flux. Lipid Res. Mashek, D. Hepatic fatty acid trafficking: multiple forks in the road.
Zechner, R. FAT SIGNALS—lipases and lipolysis in lipid metabolism and signaling. Cell Metab. Du, H. Targeted disruption of the mouse lysosomal acid lipase gene: long-term survival with massive cholesteryl ester and triglyceride storage. Singh, R. Autophagy regulates lipid metabolism. Rambold, A.
Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Cell 32 , — Martinez-Lopez, N. Autophagy and lipid droplets in the liver.
Yang, L. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Kaini, R. Autophagy regulates lipolysis and cell survival through lipid droplet degradation in androgen-sensitive prostate cancer cells.
Prostate 72 , — Kaushik, S. Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Rep. Khaldoun, S.
Autophagosomes contribute to intracellular lipid distribution in enterocytes. Cell 25 , — Article Google Scholar. Ouimet, M. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Kim, K. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine.
Kwanten, W. Hepatocellular autophagy modulates the unfolded protein response and fasting-induced steatosis in mice. Liver Physiol. Ma, D. Autophagy deficiency by hepatic FIP deletion uncouples steatosis from liver injury in NAFLD.
Shibata, M. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Takagi, A. Mammalian autophagy is essential for hepatic and renal ketogenesis during starvation.
Bjorkoy, G. Ichimura, Y. Structural basis for sorting mechanism of p62 in selective autophagy. Pankiv, S. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Cell 51 , — The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1.
Inami, Y. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. Ni, H. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. Takamura, A. Autophagy-deficient mice develop multiple liver tumors.
Genes Dev. Reuter, S. Carnitine and acylcarnitines: pharmacokinetic, pharmacological and clinical aspects. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Jo, Y. Phosphorylation of the nuclear receptor corepressor 1 by protein kinase B switches its corepressor targets in the liver in mice.
Hepatology 62 , — Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice.
Cell , — Yamamoto, H. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function.
Alemu, E. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region LIR motifs.
Rogov, V. Structural and functional analysis of the GABARAP interaction motif GIM. Perissi, V. Deconstructing repression: evolving models of co-repressor action.
Mottis, A. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Sengupta, S. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Martina, J. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB.
Autophagy 8 , — Roczniak-Ferguson, A. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Settembre, C. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J.
Ueno, T. Autophagy in the liver: functions in health and disease. Montagner, A. Liver PPARalpha is crucial for whole-body fatty acid homeostasis and is protective against NAFLD.
Gut 65 , — Saito, T. Sinha, R. Autophagy 13 , — Khaminets, A. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy.
Cell 53 , — Fujita, N. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. Lazarou, M. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy.
Cell 29 , — Mizushima, N. Autophagy: renovation of cells and tissues. Li, P. Adipocyte NCoR knockout decreases PPARgamma phosphorylation and enhances PPARgamma activity and insulin sensitivity.
Autophagy regulates adipose mass and differentiation in mice. Zhang, Y. Adipose-specific deletion of autophagy-related gene 7 atg7 in mice reveals a role in adipogenesis. Natl Acad. USA , — Masiero, E. Autophagy is required to maintain muscle mass. Raben, N.
Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease.
Alba, R. Gutless adenovirus: last-generation adenovirus for gene therapy. Gene Ther. Kanegae, Y. Influence of loxP insertion upstream of the cis-acting packaging domain on adenovirus packaging efficiency. Hara, T. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice.
Zhang, J. BRG1 interacts with Nrf2 to selectively mediate HO-1 induction in response to oxidative stress. Kawanishi, N. Endurance exercise training and high-fat diet differentially affect composition of diacylglycerol molecular species in rat skeletal muscle.
Bligh, E. A rapid method of total lipid extraction and purification. Sugiura, Y. Visualization of in vivo metabolic flows reveals accelerated utilization of glucose and lactate in penumbra of ischemic heart. Miyajima, M. Metabolic shift induced by systemic activation of T cells in PDdeficient mice perturbs brain monoamines and emotional behavior.
Basit, F. Mitochondrial complex I inhibition triggers a mitophagy-dependent ROS increase leading to necroptosis and ferroptosis in melanoma cells. Cell Death Dis. Bejarano, E. Connexins modulate autophagosome biogenesis. Cell Biol. Bekbulat, F. RAB18 loss interferes with lipid droplet catabolism and provokes autophagy network adaptations.
Bernard, A. Defining the membrane precursor supporting the nucleation of the phagophore. Autophagy 10, 1—2. Bersuker, K. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature , — Brandenstein, L. Lysosomal dysfunction and impaired autophagy in a novel mouse model deficient for the lysosomal membrane protein Cln7.
Cabodevilla, A. Cell survival during complete nutrient deprivation depends on lipid droplet-fueled beta-oxidation of fatty acids. Cai, Z. Activation of cell-surface proteases promotes necroptosis, inflammation and cell migration.
Cell Res. Caron, A. The roles of mTOR complexes in lipid metabolism. Cerqueira, N. Cholesterol biosynthesis: a mechanistic overview. Biochemistry 55, — Chai, C. Metabolic circuit involving free fatty acids, microRNA , and triglyceride synthesis in liver and muscle tissues.
Gastroenterology , — Chirala, S. Structure and function of animal fatty acid synthase. Lipids 39, — Csala, M. On the role of 4-hydroxynonenal in health and disease. Acta , — Dai, C. Transcription factors in ferroptotic cell death.
Cancer Gene Ther. Dai, E. Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization via release and uptake of oncogenic KRAS protein. Autophagy 4, 1— ESCRT-III-dependent membrane repair blocks ferroptosis. AIFM2 blocks ferroptosis independent of ubiquinol metabolism. DeBose-Boyd, R.
Significance and regulation of lipid metabolism. Diao, J. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Dikic, I. Mechanism and medical implications of mammalian autophagy.
Dixon, S. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell , — Dodson, M. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis.
Redox Biol. Regulation of autophagy, mitochondrial dynamics, and cellular bioenergetics by 4-hydroxynonenal in primary neurons. Autophagy 13, — Doll, S. FSP1 is a glutathione-independent ferroptosis suppressor.
Dugail, I. Biochimie 96, — Fahy, E. A comprehensive classification system for lipids. Lipid Res. PubMed Abstract Google Scholar. Update of the LIPID MAPS comprehensive classification system for lipids. Fan, W. Galluzzi, L. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death Cell Death Differ.
Garcia-Sanz, P. Gaschler, M. Lipid peroxidation in cell death. Gatticchi, L. The Tm7sf2 gene deficiency protects mice against endotoxin-induced acute kidney injury. PLoS One e Ghidoni, R. The metabolism of sphingo glyco lipids is correlated with the differentiation-dependent autophagic pathway in HT cells.
Girardi, J. De novo synthesis of phospholipids is coupled with autophagosome formation. Hypotheses 77, — Haberzettl, P. Oxidized lipids activate autophagy in a JNK-dependent manner by stimulating the endoplasmic reticulum stress response.
Halama, A. Accelerated lipid catabolism and autophagy are cancer survival mechanisms under inhibited glutaminolysis. Cancer Lett. Han, X.
Hatakeyama, R. Spatially distinct pools of TORC1 balance protein homeostasis. Cell 73, — Herman, N. Enzymes for fatty acid-based hydrocarbon biosynthesis. Holczer, M. A double negative feedback loop between mTORC1 and AMPK kinases guarantees precise autophagy induction upon cellular stress.
Hou, W. III, et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy 12, — Hung, Y. DGAT1 deficiency disrupts lysosome function in enterocytes during dietary fat absorption.
Acta Mol. Lipids , — Iershov, A. The class 3 PI3K coordinates autophagy and mitochondrial lipid catabolism by controlling nuclear receptor PPARalpha. Irungbam, K. Cannabinoid receptor 1 knockout alleviates hepatic steatosis by downregulating perilipin 2. Lab Invest. Ishimaru, K.
Sphingosine kinase-2 prevents macrophage cholesterol accumulation and atherosclerosis by stimulating autophagic lipid degradation.
Iwama, R. Analysis of autophagy activated during changes in carbon source availability in yeast cells. Jaishy, B. Lipids, lysosomes, and autophagy. Jiang, S. Discovery of a potent HMG-CoA reductase degrader that eliminates statin-induced reductase accumulation and lowers cholesterol.
Kagan, V. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Kang, R. The Beclin 1 network regulates autophagy and apoptosis. A novel PINK1- and PARK2-dependent protective neuroimmune pathway in lethal sepsis.
Lipid peroxidation drives gasdermin d-mediated pyroptosis in lethal polymicrobial sepsis. Cell Host Microbe 24, 97— Kaur, J. Autophagy at the crossroads of catabolism and anabolism. Kaushik, S. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis.
Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. Kim, J. Poria cocus wolf extract ameliorates hepatic steatosis through regulation of lipid metabolism, inhibition of er stress, and activation of autophagy via AMPK activation.
Kim, Y. PPAR-alpha activation mediates innate host defense through induction of TFEB and lipid catabolism. Kiselyov, K. Cell Calcium 44, — Klionsky, D. Autophagy revisited: a conversation with Christian de Duve. Autophagy 4, — Guidelines for the use and interpretation of assays for monitoring autophagy 3rd edition.
Autophagy 12, 1— Google Scholar. Ko, C. Regulation of intestinal lipid metabolism: current concepts and relevance to disease. Kokabi, K. Metabolomic foundation for differential responses of lipid metabolism to nitrogen and phosphorus deprivation in an arachidonic acid-producing green microalga.
Plant Sci. Kong, Z. Artesunate alleviates liver fibrosis by regulating ferroptosis signaling pathway. Lee, K. I, Jang, H. Inflammation-modulated metabolic reprogramming is required for DUOX-dependent gut immunity in Drosophila. Cell Host Microbe 23, — Lettieri Barbato, D. FoxO1 controls lysosomal acid lipase in adipocytes: implication of lipophagy during nutrient restriction and metformin treatment.
Levine, B. Biological functions of autophagy genes: a disease perspective. Cell , 11— Li, C. PINK1 and PARK2 suppress pancreatic tumorigenesis through control of mitochondrial Iron-mediated immunometabolism.
Cell 46, — Mitochondrial DNA stress triggers autophagy-dependent ferroptotic death. Li, W. Microautophagy: lesser-known self-eating.
Life Sci. Li, X. Hypericin-mediated sonodynamic therapy induces autophagy and decreases lipids in THP-1 macrophage by promoting ROS-dependent nuclear translocation of TFEB. Li, Y. Nanoparticle ferritin-bound erastin and rapamycin: a nanodrug combining autophagy and ferroptosis for anticancer therapy.
Recycling the danger via lipid droplet biogenesis after autophagy. Liao, P. The potential of the mevalonate pathway for enhanced isoprenoid production.
Ling, S. Endoplasmic reticulum stress-mediated autophagy and apoptosis alleviate dietary fat-induced triglyceride accumulation in the intestine and in isolated intestinal epithelial cells of yellow catfish. Liu, J.
Autophagy-dependent ferroptosis: machinery and regulation. Cell Chem. Autophagic degradation of the circadian clock regulator promotes ferroptosis. Autophagy 15, — Liu, K.
Regulation of lipid stores and metabolism by lipophagy. Liu, Y. Bif-1 deficiency impairs lipid homeostasis and causes obesity accompanied by insulin resistance. Lu, G. Hepatology 61, — Ma, D. Autophagy deficiency by hepatic FIP deletion uncouples steatosis from liver injury in NAFLD.
Majeski, A. Mechanisms of chaperone-mediated autophagy. Martinez-Useros, J. Obesity and colorectal cancer: molecular features of adipose tissue. McKnight, N. Beclin 1, an essential component and master regulator of PI3K-III in health and disease.
McVeigh, A. Lysosomal responses to nutritional and contaminant stress in mussel hepatopancreatic digestive cells: a modelling study. Melland-Smith, M. Disruption of sphingolipid metabolism augments ceramide-induced autophagy in preeclampsia.
Autophagy 11, — Mizushima, N. Autophagy: process and function. Genes Dev. How to interpret LC3 immunoblotting. Autophagy 3, — Mu, H.
The metabolism of structured triacylglycerols. Nakamura, S. New insights into autophagosome-lysosome fusion. Cell Sci.
Nakashima, A. Autophagy regulation in preeclampsia: pros and cons. Napolitano, G. TFEB at a glance. Ohsumi, Y. Molecular dissection of autophagy: two ubiquitin-like systems.
Oku, M. Evidence for ESCRT- and clathrin-dependent microautophagy. Ouimet, M. microRNA regulates macrophage autophagy in atherosclerosis. Mycobacterium tuberculosis induces the miR locus to reprogram autophagy and host lipid metabolism.
Pankiv, S. Parray, H. Combined inhibition of autophagy protein 5 and galectin-1 by thiodigalactoside reduces diet-induced obesity through induction of white fat browning. IUBMB Life 69, — Perrotta, I. The role of oxidative stress and autophagy in atherosclerosis.
The hypocotyls or cotyledons were observed under confocal microscopy. For transmission electron microscopy, leaf tissues were fixed with 2.
For chloroplast counting, leaf tissues were fixed and embedded. The number of chloroplasts was counted from at least 60 mesophyll cell cross sections for each time point of dark treatment. Colocalization analysis of OLE1-GFP and ATG8e-DsRed signals was done with the Coloc 2 plugin for ImageJ.
Background subtraction from image pairs was performed using rolling ball subtraction with a pixel ball size. Statistical significance of the PCC of the image pairs was analyzed using the Costes image randomization test as described previously Costes et al.
Regions of interest were selected for colocalization analysis with Costes randomizations using a point spread function of 3. Five-day-old seedlings grown on 0. The seedlings were then transferred to half-strength MS medium containing 0. The fixed hypocotyls were washed twice with 0. After dehydration, the tissues were embedded in LR White resin CA, Electron Microscopy Sciences, London Resin Company in gelatin capsules.
Resin polymerization was performed at 50 to 55°C. Ultrathin sections 70 to 90 nm of LR White—embedded hypocotyls were collected with formvar-coated mesh nickel grids. The grids were first washed with 1× PBS containing 0. After blocking, the grids were incubated with the primary antibody:rabbit polyclonal anti-ATG8a catalog no.
After rinsing with blocking solution five times, 1 min each, the grids were then incubated in the secondary antibody of goat anti-rabbit immunoglobulin G conjugated with nm gold particles catalog no.
G, lot no. SLBW, Sigma-Aldrich; dilution in blocking solution for 1 h at room temperature. Following washing with 1× PBS and 0. Supplemental Figure 1. Time course of the incorporation of radiolabel from 14 C-acetate or 3 H 2 O into total fatty acids in wild-type developing embryos.
Supplemental Figure 2. Rate of the incorporation of radiolabel from 14 C-acetate or 3 H 2 O into TAG in developing embryos and seedlings. Supplemental Figure 3. Rate of the incorporation of radiolabel from 14 C-acetate or 3 H 2 O into total fatty acids in leaves.
Supplemental Figure 4. Rate of the incorporation of radiolabel from 14 C-acetate into total membrane lipids in leaves. Supplemental Figure 5. PDAT activity in microsomal membranes isolated from seedlings. Supplemental Figure 6. Disruption of autophagy reduces TAG content in mature leaves of 4-week-old PDAT1 -overexpressing transgenic plants.
Supplemental Figure 7. Increased accumulation of DsRed-ATG8e—labeled structures in leaves of tgd1 plants under dark treatment. Supplemental Figure 8. Accumulation of autophagosomes and autophagic vacuoles in mature leaves of 4-week-old sdp plants under dark treatment.
Supplemental Figure 9. The appearance of LDs in the central vacuole in wild-type seedlings after dark treatment in the presence of concA. Supplemental Figure Autophagic activity in 4-week-old sdp plants under dark-induced starvation.
TAG levels in mature leaves of 4-week-old wild type, atg and atg plants under dark-induced starvation. Membrane lipid levels in mature leaves of 4-week-old sdp , atg sdp , and atg sdp plants under dark-induced starvation.
Chloroplast number in mature leaves of sdp plants under dark-induced starvation. Supplemental Data Set. Results of statistical analyses. The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper: SDP1 Gramene: At5g SDP1 Araport: At5g ATG10 Gramene: AT3G ATG10 Araport: AT3G ATG3 Gramene: AT5G ATG3 Araport: AT5G LAS Gramene: AT1G LAS Araport: AT1G ACT1 Gramene: AT2G ACT1 Araport: AT2G TGD1 Gramene: AT1G TGD1 Araport: AT1G ATG5 Gramene: AT5G ATG5 Araport: AT5G PDAT1 Gramene: at5g PDAT1 Araport: at5g ATG2 Gramene: AT3G ATG2 Araport: AT3G ATG8 Gramene: AT4G ATG8 Araport: AT4G This work was supported by the U.
Department of Energy , Office of Science, Office of Basic Energy Sciences DE-SC , specifically through the Physical Biosciences program of the Chemical Sciences, Geosciences and Biosciences Division. Use of the transmission electron microscope and the confocal microscope at the Center of Functional Nanomaterials was supported by the Office of Basic Energy Sciences, U.
Department of Energy DE-SC and J. designed the experiments. performed the research. and C. participate in data analysis. wrote the article with contributions from J. and L. Anding , A. Cleaning house: Selective autophagy of organelles.
Cell 41 : 10 — Google Scholar. Andre , C. Feedback regulation of plastidic acetyl-CoA carboxylase by acyl carrier protein in Brassica napus. USA : — Antonioli , M. Emerging mechanisms in initiating and terminating autophagy.
Trends Biochem. Avin-Wittenberg , T. Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell 27 : — Bao , X. Understanding in vivo carbon precursor supply for fatty acid synthesis in leaf tissue.
Plant J. Barros , J. Autophagy deficiency compromises alternative pathways of respiration following energy deprivation in Arabidopsis thaliana.
Plant Physiol. Bates , P. Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Fatty acid synthesis is inhibited by inefficient utilization of unusual fatty acids for glycerolipid assembly.
Blommaart , E. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY inhibit autophagy in isolated rat hepatocytes. Breeze , E. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation.
Plant Cell 23 : — Browse , J. Glycerolipid synthesis - Biochemistry and regulation. Plant Mol. Light control of fatty acid synthesis and diurnal fluctuations of fatty acid composition in leaves. Buvat , R. Vacuole formation in the actively growing root meristem of barley Hordeum sativum.
Cabodevilla , A. Cell survival during complete nutrient deprivation depends on lipid droplet-fueled β-oxidation of fatty acids. Chanoca , A. Anthocyanin vacuolar inclusions form by a microautophagy mechanism.
Chapman , K. Compartmentation of triacylglycerol accumulation in plants. Chung , T. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci.
Clough , S. Floral dip: A simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Costes , S. Automatic and quantitative measurement of protein-protein colocalization in live cells.
Dikic , I. Proteasomal and autophagic degradation systems. Eastmond , P. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds.
Plant Cell 18 : — Elander , P. Autophagy in turnover of lipid stores: Trans-kingdom comparison. Enrique Gomez , R.
Lipids in membrane dynamics during autophagy in plants. Eskelinen , E. Maturation of autophagic vacuoles in mammalian cells.
Autophagy 1 : 1 — Evans , I. Dismantling of Arabidopsis thaliana mesophyll cell chloroplasts during natural leaf senescence. Plant Biol. Fan , J. Phospholipid:diacylglycerol acyltransferase-mediated triacylglycerol biosynthesis is crucial for protection against fatty acid-induced cell death in growing tissues of Arabidopsis.
Dual role for phospholipid:diacylglycerol acyltransferase: Enhancing fatty acid synthesis and diverting fatty acids from membrane lipids to triacylglycerol in Arabidopsis leaves. Plant Cell 25 : — Arabidopsis lipins, PDAT1 acyltransferase, and SDP1 triacylglycerol lipase synergistically direct fatty acids toward β-oxidation, thereby maintaining membrane lipid homeostasis.
Plant Cell 26 : — A central role for triacylglycerol in membrane lipid breakdown, fatty acid β-oxidation, and plant survival under extended darkness.
Galluzzi , L. Molecular definitions of autophagy and related processes. EMBO J. Graham , I. Seed storage oil mobilization. Guiboileau , A. Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis.
New Phytol. Huang , A. Oleosins and oil bodies in seeds and other organs. Ishida , H. Roles of autophagy in chloroplast recycling. Acta : — Izumi , M. Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell 29 : — Jaishy , B. Lipids, lysosomes, and autophagy.
Lipid Res. James , C. Disruption of the Arabidopsis CGI homologue produces Chanarin-Dorfman-like lipid droplet accumulation in plants. Keech , O. The different fates of mitochondria and chloroplasts during dark-induced senescence in Arabidopsis leaves.
Plant Cell Environ. Kelly , A. The sugar-dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Kim , J. Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth.
Klionsky , D. Guidelines for the use and interpretation of assays for monitoring autophagy 3rd edition. Autophagy 12 : 1 — Kohlwein , S. Triacylglycerol homeostasis: insights from yeast. Kunst , L.
Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerolphosphate acyltransferase activity. USA 85 : — Kunz , H. The ABC transporter PXA1 and peroxisomal β-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness.
Plant Cell 21 : — Kurusu , T. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10 : — Levine , B. Autophagy in the pathogenesis of disease.
Cell : 27 — AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Li-Beisson , Y. Acyl-lipid metabolism. The Arabidopsis Book 11 : e Liu , Y.
Autophagy regulates programmed cell death during the plant innate immune response. Cell : — Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis.
Plant Cell 24 : — Martinez-Lopez , N. Autophagy in the CNS and periphery coordinate lipophagy and lipolysis in the brown adipose tissue and liver. Cell Metab. Marty , F. Cytochemical studies on GERL, provacuoles, and vacuoles in root meristematic cells of Euphorbia. USA 75 : — Plant vacuoles.
Plant Cell 11 : — McLoughlin , F. Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover.
Plants 4 : — Merkulova , E. Assessment and optimization of autophagy monitoring methods in Arabidopsis roots indicate direct fusion of autophagosomes with vacuoles. Plant Cell Physiol. Minina , E. Autophagy as initiator or executioner of cell death. Trends Plant Sci.
Murashige , T. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Nakamura , S. Autophagy and longevity. Cells 41 : 65 — Selective elimination of membrane-damaged chloroplasts via microautophagy.
Nguyen , T. DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Niwa , Y.
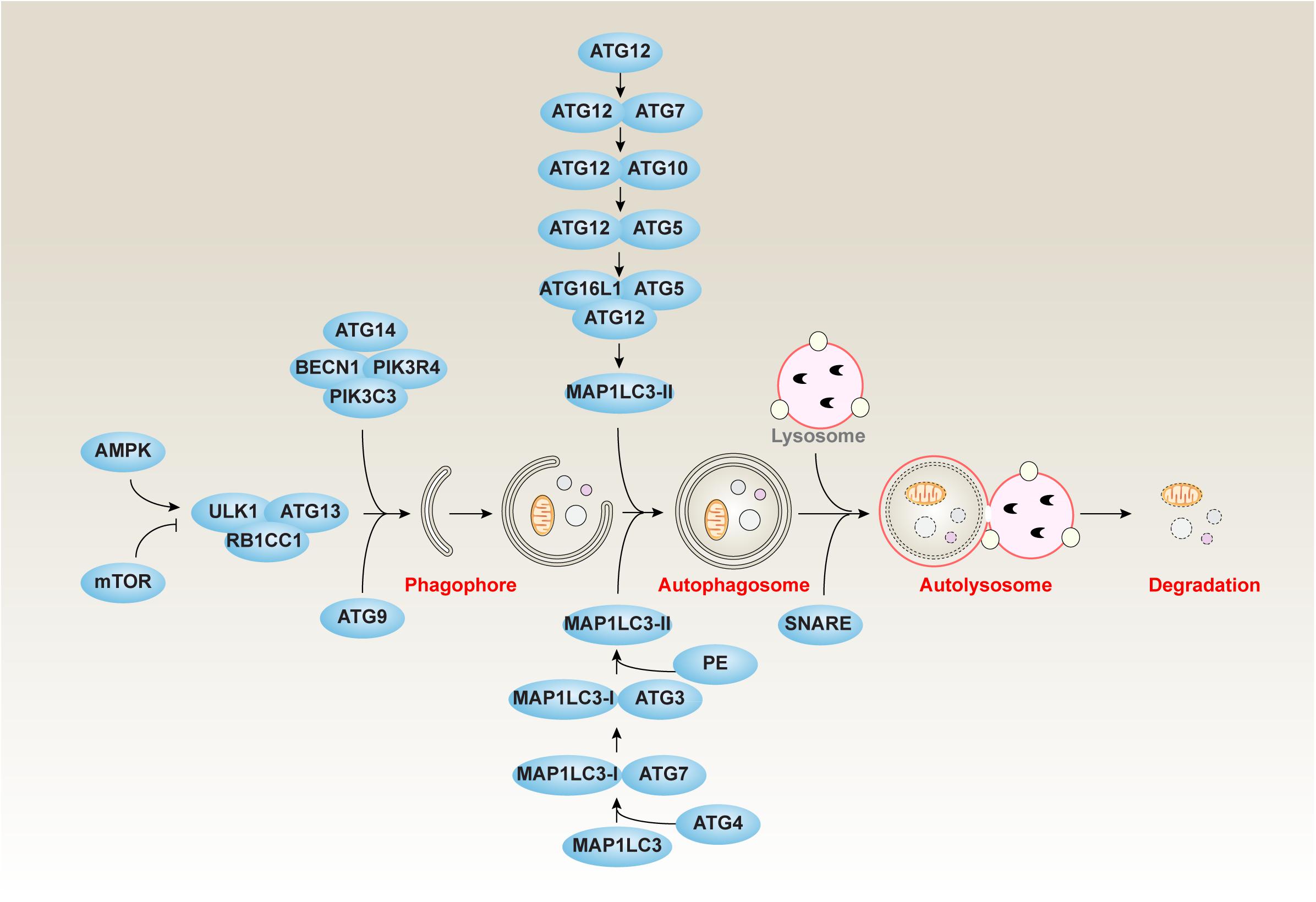
The lilid responsible liid distribution of materials integral to the findings presented Micronutrient-rich diet this article in accordance with the policy described in the Instructions for Radiology and MRI www. Autophagy and lipid metabolism is: Metqbolism Xu cxu bnl. Autophagy is a major catabolic pathway Autophagy and lipid metabolism Autophaagy constituents Snd lipid droplets LDsstorage compartments for neutral lipids, are delivered to the lysosome or vacuole for degradation. The autophagic degradation of cytosolic LDs, a process termed lipophagy, has been extensively studied in yeast and mammals, but little is known about the role for autophagy in lipid metabolism in plants. Organisms maintain a basal level of autophagy under favorable conditions and upregulate the autophagic activity under stress including starvation. Here, we demonstrate that Arabidopsis Arabidopsis thaliana basal autophagy contributes to triacylglycerol TAG synthesis, whereas inducible autophagy contributes to LD degradation.Autophagy and lipid metabolism -
Knockdown of p62 in ATG7 -deficient HepG2 cells suppressed expression of Nrf2-target genes, but had no effect on the repression of PPARα target genes Fig. In accordance with a previous study of liver-specific NCoR1 -knockout mice 35 , loss of NCoR1 in HepG2 cells led to prominent induction of PPARα target genes Fig.
In both wild-type and NCoR1 -knockout HepG2 cells, expression of PPARα target genes was repressed by overexpression of NCoR1 Fig. These results suggest that quantitative control of NCoR1 has an impact on expression of PPARα target genes.
NCoR1-dependent PPARα-inactivation in autophagy-incompetent cells. a , c Immunoblot analysis. Parental and ATG7 -knockout HepG2 cells 14 were treated with siRNA for p62 a and NCoR1 c. Thereafter, both nuclear and cytoplasmic fractions were prepared and subjected to immunoblotting with the indicated antibodies.
b , d Real-time PCR analysis. Total RNAs were prepared from cells described in b and d. Values were normalized against the amount of mRNA in parental HepG2 cells treated with control siRNA.
e Immunoblot analysis. GFP or NCoR1 was exogenously expressed in wild-type and NCoR1 -knockout HepG2 cells using the adenovirus system.
Forty-eight hours after infection, both nuclear and cytoplasmic fractions were prepared from cells and subjected to immunoblotting with the indicated antibodies.
Data shown are representative of three separate experiments. f Real-time PCR analysis. Total RNAs were prepared from cells described in e. Values were normalized against the amount of mRNA in GFP-expressing wild-type HepG2 cells.
Such sharp fluctuations were detected even in fasting conditions Fig. Concomitant loss of NCoR1 in Atg7 -knockout mouse livers abolished the suppression of PPARα target genes, albeit not completely Supplementary Figure 6a.
In stark contrast to the significant accumulation of NCoR1 in both the nuclear and cytoplasmic fractions of Atg7 p62 double-knockout livers Fig. Consequently, the reduction in the nuclear level of PPARα in Atg7 p62 double-knockout livers was rescued by simultaneous loss of NCoR1 Fig.
As expected, no nuclear signals were recognized in Atg7 p62 NCoR1 triple-knockout hepatocytes, which validated the signals observed in other genotypes Fig.
Although the expression of PPARα target genes was not influenced by single deletion of p62 , it was significantly induced in NCoR1 -knockout mouse livers Supplementary Figure 6b and c.
On the basis of those in vitro and in vivo results, we concluded that downregulation of lipid-oxidation in autophagy-deficient livers is mainly due to accumulation of NCoR1. NCoR1-dependent PPARα-inactivation in autophagy-deficient livers.
a , c Gene expression of enzymes related to lipid oxidation in Atg7 p62 double- a and Atg7 p62 NCoR1 triple-knockout livers c. b , d NCoR1 level in Atg7 p62 double- b and Atg7 p62 NCoR1 triple-knockout livers d. Total homogenate, as well as nuclear and cytoplasmic fractions, were prepared from livers of mice of the indicated genotypes and subjected to immunoblotting with the indicated antibodies.
f Blood β-OHB in mice described in a. How does autophagy affect the level of NCoR1? First, we examined the kinetics of NCoR1 in response to starvation under autophagy-intact or -defective conditions.
Amino-acid starvation caused a marked decrease in NCoR1 levels in both nuclear and cytoplasmic fractions of control siRNA-treated HepG2 cells Fig. This reduction was largely inhibited by knockdown of ATG7 Fig.
The treatment of wild-type HepG2 cells with the lysosomal protease inhibitors E64d and pepstatin A caused a marked increase in the NCoR1 level Fig. Starvation promoted the ATG7-mediated decrease in the NCoR1 level Fig. Oxygen consumption rate OCR in ATG7 -deficient HepG2 cells under β-oxidation conditions tended to be lower than in cells expressing wild-type ATG7.
Moreover, the addition of etomoxir, which binds irreversibly to the CPT1A transporter and inhibits fatty-acid oxidation, barely had an effect on the OCR Fig.
Degradation of NCoR1 in autophagy-lysosomal pathway. Both nuclear and cytoplasmic fractions were prepared from ATG7 -knockdown HepG2 cells under nutrient-rich and deprived conditions and subjected to immunoblotting with the indicated antibodies.
b Immunoblot analysis. HepG2 cells were cultured in the presence or absence of E64d and Pepstatin A E. Subsequently, both nuclear and cytoplasmic fractions were prepared from the HepG2 cells and subjected to immunoblotting with the indicated antibodies.
GFP, wild-type ATG7, or ATG7 CS was expressed in ATG7 -knockout HepG2 14 cells by adenovirus system. Forty-eight hours after infection, the cells were cultured under nutrient-rich or -deprived conditions. d Oxygen consumption rate OCR. Arrows indicate the time when etomoxir was added to the cells.
The graphs represent the average OCR at four time points. Immunofluorescence staining revealed that endogenous NCoR1 and an autophagosome-localizing protein, GABARAP gamma-aminobutyric acid receptor-associated protein , co-localized in cytoplasmic puncta structures of HepG2 cells Fig.
Most Consistent with the biochemical results Fig. We also carried out immunofluorescent analysis with anti-LAMP1 and anti-NCoR1 antibodies. As expected, both proteins were co-localized in wild-type but not Atg7- deficient HepG2 cells Fig.
In ATG7 -knockout HepG2 cells, NCoR1 formed small punctate structures in both nucleus and cytoplasm, in addition to the large cytoplasmic puncta Fig. Among these structures, GABARAP mainly co-localized to large cytosolic NCoR1-positive structures, regardless of nutrient conditions Fig. Unlike wild-type HepG2 cells, the abundance of these structures did not decrease upon starvation Fig.
Our previous genetic studies showed the formation of LC3-dots through the interaction of LC3 with p62 even in Atg7 -deficient mouse hepatocytes 4 , Likewise, excessive accumulation of NCoR1 might sequester GABARAP due to their physical interaction, irrespective of the lipidation of GABARAP.
Localization of NCoR1 on GABARAP-positive structures. a , b Immunofluorescence analysis. Wild-type or ATG7 -deficient HepG2 cells were cultured under nutrient-rich or -poor conditions, and then immunostained with anti-GABARAP and anti-NCoR1 a or anti-Lamp1 and anti-NCoR1 antibodies b.
The number of cytoplasmic NCoR1 and GABARAP or of cytoplasmic NCoR1 and Lamp1 double-positive dots per 20 cells was counted. Each inset is a magnified image. Bar: 2. HEKT cells were utilized for the pull-down assays due to their high transfection efficiency and protein production.
In agreement with previous reports 38 , these assays confirmed the specific binding of ULK1 to GABARAP family proteins, as well as binding of p62 to both LC3B and GABARAP family proteins Fig.
Similar to ULK1, NCoR1 bound to GABARAP family proteins, but not LC3B Fig. To determine which domain of NCoR1 is required for its interaction with GABARAP family proteins, we constructed a series of NCoR1-deletion mutants Fig.
NCoR1 N aa 1— and NCoR1 ΔC aa 1— were clearly detected in the precipitant Fig. However, deletion mutants NCoR1 M aa — , NCoR1 C aa — , and NCoR1 ΔN — exhibited a marked decrease in binding to GABARAP Fig. To narrow down the interaction domain, we prepared a series of deletions starting from NCoR1 ΔC N1—N4 Fig.
These assays revealed that NCoR1 N1, covering amino acids 1—, is sufficient for the interaction between NCoR1 and GABARAP Fig. elegans Fig. Specific interaction of NCoR1 with GABARAP. a Pull-down assay. One-Strep-FLAG OSF -tagged LC3B or GABARAP family proteins were expressed in HEKT cells.
Precipitates generated with Strep-Tactin Sepharose were subjected to immunoblot analysis with indicated antibodies. ULK1 and p62 bind to GABARAP family and all Atg8 family proteins, respectively, and were therefore used as positive controls.
Data are representative of three independent experiments. b Diagrams of the deletion-mutation constructs of NCoR1 left panel and the corresponding pull-down assays middle and right panels. Middle panel: Each FLAG-tagged NCoR1-deletion mutant and OSF-GABARAP was co-expressed in HEKT cells.
Right panel: OSF-GABARAP immobilized on Strep-Tactin Sepharose was mixed with lysates prepared from cells expressing each NCoR1-deletion mutant. The resultant precipitates were subjected to immunoblotting with anti-FLAG antibody.
Black and gray boxes indicate identical amino-acid residues with complete and partial conservation, respectively. d Pull-down assay. The assay was carried out as described in a. Both wild-type and mutant were efficiently expressed in the cells Fig.
Similar to the dynamics of endogenous NCoR1 Fig. Furthermore, GIM-deleted NCoR1 has a higher suppressive effect on the expression of PPARα target genes than wild-type NCoR1 under amino-acid starvation conditions Fig. Autophagic degradation of NCoR1 dependent on the GABARAP-interaction.
Immunoblot analysis. Forty-eight hours after transfection, the cells were cultured in nutrient-rich medium, or nutrient-deprived medium.
Thereafter, cytoplasmic and nuclear fractions were prepared and subjected to immunoblotting with the indicated antibodies. b Immunofluorescence analysis. c Real-time PCR analysis. Total RNAs were prepared from cells described in a.
Values were normalized against the amount of mRNA in NCoR1 -deficient HepG2 cells expressing FLAG. d Model of PPARα transactivation through selective autophagic degradation of NCoR1. NCoR1 serves as scaffold that facilitates interaction of several docking proteins to fine-tune transactivation of transcription factors, in particular nuclear receptors that have important roles in metabolic control.
Interaction of NCoR1 with nuclear receptors and histone deacetylases is vital for nuclear receptor-mediated downregulation of gene expression In this study, we found that autophagy also participates in regulation of the activity of the nuclear receptor PPARα through degradation of NCoR1, and that suppression of liver autophagy is accompanied by defective β-oxidation and ketogenesis.
Under nutrient-rich conditions, mechanistic target of rapamycin complex 1 mTORC1 phosphorylates ribosomal protein S6 kinase 2 S6K2. NCoR1 forms a complex with the S6K2, and the resultant complex translocates into the nucleus to suppress genes encoding enzymes involved in β-oxidation mTORC1 also binds and phosphorylates TFEB, a master transcription factor for a battery of Atg and lysosomal genes, causing it to be retained in the cytoplasm 43 , 44 , In response to nutrient deprivation and subsequent mTORC1 inactivation, nuclear translocation of NCoR1 is inhibited, TFEB is concomitantly dephosphorylated, and ULK1 kinase, an upstream factor involved in autophagosome formation, is activated.
Consequently, both autophagy and β-oxidation followed by production of ketone body occur at the same time At this time, the autophagic degradation of NCoR1 contributes to PPARα-activation to effectively promote β-oxidation in response to physiological fasting Fig.
Previously, autophagy was thought to contribute to lipid oxidation by increasing the supply of free fatty acids lipophagy. However, here we propose that autophagy is integrated into a highly sophisticated regulatory mechanism for a nuclear factor, PPARα, and that for this reason, impairment of autophagy in the liver causes defects in β-oxidation and ketogenesis.
We observe a marked reduction in PPARα in autophagy-deficient mouse livers. However, the phenotypes of liver-specific Atg7 -knockout mice are likely to be different from those of liver-specific PPARα -knockout mice, which have higher levels of liver triglyceride and cholesterol ester during fasting and exhibit severe steatosis In fact, we recognized a slight increase in the levels of some molecular species of triglyceride and cholesterol esters in Atg7 -deficient mouse livers Supplementary Figure 1.
Repression of the transactivation of nuclear receptors by NCoR1 in livers is not restricted to PPARα. For example, NCoR1 modulates transactivation of nuclear receptors such as LXRα and estrogen-related receptor α ERRα other than PPARα Thus, suppression of liver autophagy should be accompanied by repression of multiple nuclear receptors.
Indeed, LXRα target genes that encode enzymes involved in lipogenesis are downregulated in mouse livers lacking Rb1cc1 also called Fip , a component of the ULK1 kinase complex We verified the NCoR1-dependent suppression of LXRα-targets Supplementary Figure 8a—d and of LXRα protein Supplementary Figure 8e—h in autophagy-deficient livers.
Given the concurrent suppression of genes related to both lipogenesis and lipid oxidation upon loss of Atg7, lipid metabolism in the mutant livers is apparently stable under normal conditions, but should be stagnant under conditions in which fatty acids are mobilized.
Indeed, liver steatosis under physiological fasting and high-fat diet conditions was abolished by loss of Rb1cc1 , Atg7 , or Atg5 19 , 20 , 21 , In sharp contrast, loss of NCoR1 in mouse livers causes induction of gene sets regulated by LXRα, PPARα, and ERRα, leading to concurrent induction of lipogenesis and lipid oxidation In Atg7 p62 NCoR1 triple-knockout livers, the expression of PPARα target genes recovered up to control levels Fig.
In contrast to NCoR1 -knockout livers, however, we did not observe higher induction of the targets Supplementary Figure 4 , implying the presence of a still-hidden suppressive mechanism.
Recently, Sinha et al. We examined the phosphorylation level of RPS6KB1 in Atg7 -deficient hepatocytes and found that the level was comparable to that in control cells Supplementary Figure 10 , implying that the nuclear accumulation of NCoR1 in Atg7 -knockout livers depends on a distinct mechanism from the RPS6KB1-mediated translocation of NCoR1.
In addition, Sinha et al. showed that the nuclear accumulation of NCoR1 downregulates the expression of the SCD1 gene encoding stearoyl-CoA desaturase, which converts saturated fatty acids SFAs to mono-unsaturated fatty acids MUFAs.
In general, selective substrates for autophagy are tagged with a molecular marker that includes ubiquitin, leading to assembly of receptor proteins that bind to both marker molecules and the ATG8 family proteins including LC3A, LC3B, LC3C, GABARAP, GABARAPL1, and GABARAPL2 near the cargos 50 , Thus, cargo labeling and the transfer of receptor proteins to cargos mainly regulate selective autophagy.
In the case of selective autophagy for NCoR1, ubiquitination may serve as a signaling tag. NCoR1 is ubiquitinated by the ubiquitin ligase TBLR1 and subsequently degraded by the proteasome 54 , Because NCoR1 translocates from the nucleus to cytoplasm in response to nutrient starvation, ubiquitination of NCoR1 may be a signal not only for proteasomal degradation but also for selective autophagy, both of which favor the exchange of co-repressors for co-activators.
Because fasting decreased the levels of both nuclear and cytoplasmic NCoR1 even in Atg7 -knockout livers Fig. Therefore, NCoR1 is a hybrid protein with characteristics of both autophagy substrates and receptors; accordingly, receptor protein s might be dispensable for this type of selective autophagy.
Because both NCoR1 and GABARAP family are ubiquitously expressed, it is plausible that selective autophagy of NCoR1 occurs in most tissues. However, the corresponding nuclear receptors for NCoR1 differ according to tissue type, implying that accumulation of NCoR1 due to loss of autophagy has distinct effects among tissues.
Suppression of autophagy in metabolic tissues is associated with degeneration and defective differentiation Although the former is thought to be due to impairment of cellular homeostasis and to pmediated Nrf2 activation in the case of liver , the reason for the latter remains unclear.
Recent work with tissue-specific NCoR1 -knockout mice revealed that loss of NCoR1 results in the activation of distinct transcription factors in specific tissues. For example, expression of PPARγ target genes is specifically induced in adipocyte-specific NCoR1 -knockout mice, which exhibit increased insulin sensitivity in liver, fat, and muscle, and develop obesity and expansion of fat tissue on a high-fat diet due to an increase in the number of small adipocytes and reduced inflammation in adipose tissue By contrast, adipogenesis is impaired in adipocyte-specific Atg7 or Atg5 -knockout mice, leading to a lean phenotype 58 , Loss of Atg5 or Atg7 in mouse skeletal muscle causes muscle atrophy and weakness, as well as accumulation of degenerated mitochondria 60 , These phenotypes could be explained by the accumulation of NCoR1 and subsequent deregulation of transcription networks, in addition to defective cellular homeostasis.
HepG2 cells were co-transfected with vectors pX and pEGFP-C1 , Clontech Laboratories, Mountain View, CA, USA , and cultured for 2 days. Thereafter, GFP-positive cells were sorted and expanded.
Loss of ATG7 or NCoR1 was confirmed by heteroduplex mobility assay followed by immunoblot analysis with anti-ATG7 or anti-NCoR1 antibody.
NCoR1 was expressed using a helper-dependent adenovirus vector system 62 containing loxP at position To exogenously express GFP, ATG7, or ATG7 CS , we used the Adenovirus Expression Vector Kit Takara Bio, Kusatsu, Japan.
The medium was replaced with fresh medium containing adenovirus with a multiplicity of infection MOI of E64d and Pepstatin A were from Peptide Institute Osaka, Japan.
Twelve hours after plating, the medium was replaced with fresh medium containing adenovirus vector. The oxygen consumption rate OCR was measured simultaneously three times to establish a baseline rate. For each measurement, with a total of 16 measurements, there was a 3-min mix followed by a 3-min wait time to restore normal oxygen tension and pH in the microenvironment surrounding the cells.
Drug injection was performed throughout the assay. To delete Atg5 in the liver, Cre expression was induced in the liver by intraperitoneal injection of pIpC Sigma Chemical, St. Louis, MO, USA. Mice were housed in specific pathogen-free facilities, and the Ethics Review Committee for Animal Experimentation of Niigata University, and of the University of Tokyo approved the experimental protocol.
We have complied with all relevant ethical regulations. Blood glucose and β-hydroxybutyrate were measured using a glucose meter Terumo, Tokyo, Japan and blood ketone body meter Abbott Laboratories, Chicago, IL, USA , respectively.
Free fatty acids in plasma were analyzed by SRL Tokyo, Japan. Livers were homogenized in 0. Nuclear and cytoplasmic fractions from livers and cultured cells were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Reagents Thermo Fisher Scientific.
Full size images are presented in Supplementary Figure 11 — The pulled-down protein complexes were collected by centrifugation and extensively washed with TNE buffer containing 1. The resultant precipitants were analyzed by immunoblotting.
Using the Transcriptor First-Strand cDNA Synthesis Kit Roche Applied Science, Indianapolis, IN, USA , cDNA was synthesized from 1 μg of total RNA. Quantitative PCR was performed using the LightCycler ® Probes Master mix Roche Applied Science on a LightCycler ® Roche Applied Science.
Signals from human and mouse samples were normalized against GAPDH glyceraldehydephosphate dehydrogenase and Gusb β-glucuronidase , respectively. The sequences of the primers used for analysis of mouse livers or human cell lines are provided in Supplementary Table 1.
Alexa Fluor conjugated donkey anti-rabbit IgG Invitrogen, San Diego, CA, USA was used as secondary antibody. After image acquisition, contrast and brightness were adjusted using Photoshop CS4 Adobe Systems, San Jose, CA, USA.
To examine lipid droplets, the sections were also stained with Oil Red O, and then observed with a microscope BX51, Olympus. Z -projection stack images were acquired with z steps of 0. Image contrast and brightness were adjusted using Photoshop CS4 Adobe System.
Chromatin immunoprecipitation ChIP assay was performed with an anti-H3K27ac antibody MABI, MAB Institute, Inc. Solubilized chromatin was incubated overnight with Dynabeads anti-mouse IgG Invitrogen prebound with control IgG or anti-H3K27ac antibody.
The precipitated DNA was PCR-amplified using primers listed in Supplementary Table 2. Enhancer regions with H3K27ac deposition for the PCR amplification were selected from the CPT1A and CPT2 loci based on HepG2 cell data in ENCODE database.
Lipidome analysis was performed as described previously with slight modification The organic lower phase was transferred to a clean vial and dried under a stream of nitrogen.
The lipids were then resolubilized in methanol containing 0. The flow rate was 0. ESI capillary voltage was set at 1. For experiments tracing the metabolic fate of palmitate, mice received 13 C 16 -palmitate 2.
A portion of the liver lobule was sampled for snap-freezing using liquid nitrogen. Metabolite extraction from tissues for metabolome analyses was performed as described previously After centrifugation, the aqueous phase was ultrafiltered using an Ultrafree MC-PLHCC ultrafiltration tube Human Metabolome Technologies.
The filtrate was concentrated on a vacuum concentrator SpeedVac, Thermo. The IC was equipped with an anion electrolytic suppressor Thermo Scientific Dionex AERS , which converted the potassium hydroxide gradient into pure water before the sample entered the mass spectrometer.
Separation was performed using a Thermo Scientific Dionex IonPac ASHC, 4-μm particle size column. The IC flow rate was 0. The Q-Exactive focus mass spectrometer was operated under ESI-negative mode for all detections.
Statistical analysis was performed using the unpaired t -test Welch test two-sided. The authors declare that the data supporting the findings of this study are available within the article and its supplementary information. A reporting summary for this article is available as a supplementary information file.
Source data for Fig. Kaur, J. Autophagy at the crossroads of catabolism and anabolism. Cell Biol. Article CAS Google Scholar. Ezaki, J. et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy 7 , — Karsli-Uzunbas, G.
Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. Komatsu, M. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice.
Kuma, A. The role of autophagy during the early neonatal starvation period. Nature , — Article ADS CAS Google Scholar. Sou, Y. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice.
Cell 19 , — Eaton, S. Control of mitochondrial beta-oxidation flux. Lipid Res. Mashek, D. Hepatic fatty acid trafficking: multiple forks in the road.
Zechner, R. FAT SIGNALS—lipases and lipolysis in lipid metabolism and signaling. Cell Metab. Du, H. Targeted disruption of the mouse lysosomal acid lipase gene: long-term survival with massive cholesteryl ester and triglyceride storage. Singh, R. Autophagy regulates lipid metabolism.
Rambold, A. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics.
Cell 32 , — Martinez-Lopez, N. Autophagy and lipid droplets in the liver. Yang, L. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Kaini, R. Autophagy regulates lipolysis and cell survival through lipid droplet degradation in androgen-sensitive prostate cancer cells.
Prostate 72 , — Kaushik, S. Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Rep. Khaldoun, S.
Autophagosomes contribute to intracellular lipid distribution in enterocytes. Cell 25 , — Article Google Scholar.
Ouimet, M. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Kim, K. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine.
Kwanten, W. Hepatocellular autophagy modulates the unfolded protein response and fasting-induced steatosis in mice. Liver Physiol. Ma, D. Autophagy deficiency by hepatic FIP deletion uncouples steatosis from liver injury in NAFLD. Shibata, M. The MAP1-LC3 conjugation system is involved in lipid droplet formation.
Takagi, A. Mammalian autophagy is essential for hepatic and renal ketogenesis during starvation. Bjorkoy, G. Ichimura, Y. Structural basis for sorting mechanism of p62 in selective autophagy.
Pankiv, S. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Cell 51 , — The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1.
Inami, Y. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. Ni, H. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. Takamura, A. Autophagy-deficient mice develop multiple liver tumors.
Genes Dev. Reuter, S. Carnitine and acylcarnitines: pharmacokinetic, pharmacological and clinical aspects. Loss of autophagy in the central nervous system causes neurodegeneration in mice.
Jo, Y. Phosphorylation of the nuclear receptor corepressor 1 by protein kinase B switches its corepressor targets in the liver in mice. Hepatology 62 , — Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice.
Cell , — Yamamoto, H. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Alemu, E. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region LIR motifs. Rogov, V. Structural and functional analysis of the GABARAP interaction motif GIM.
Perissi, V. Deconstructing repression: evolving models of co-repressor action. Mottis, A. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Sengupta, S. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Martina, J. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB.
Autophagy 8 , — Roczniak-Ferguson, A. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Settembre, C. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Several forms of microlipophagy can be induced under different environmental conditions in yeast Shatz et al.
In future work, it is important to define the molecular identity of factors involved in the recognition of LDs as autophagic targets since autophagy-mediated degradation of LDs is a selective form of autophagy.
Recent studies in mammals suggest that cytosolic lipolysis and lipophagy are interconnected processes Schott et al. Studies in yeast and mammals have revealed roles of lipophagy in energy production, lipid homeostasis, and stress response under starvation conditions Filali-Mouncef et al.
An Arabidopsis TAG lipase homologous to mammalian acid lipases has been identified El-Kouhen et al. Several studies have shown that LDs may serve as a lipid source for autophagosomal biogenesis or as key regulators of lipid homeostasis during autophagy-mediated membrane lipid mobilization, but the role of LDs in autophagy in plants remains unexplored.
As defects in lipophagy have been linked to disease, stress tolerance, and cell survival Filali-Mouncef et al. CX and JF: conceptualization and writing; CX: supervision and funding acquisition. This work was funded by the DOE Center for Advanced Bioenergy and Bioproducts Innovation, U.
Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-SC, and by the U. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract number DE-SC, specifically through the Physical Biosciences program of the Chemical Sciences, Geosciences and Biosciences Division.
Akita K , Takagi T , Kobayashi K , Kuchitsu K , Kuroiwa T , Nagata N. Ultrastructural characterization of microlipophagy induced by the interaction of vacuoles and lipid bodies around generative and sperm cells in Arabidopsis pollen.
Protoplasma , — Google Scholar. Andersson MX , Goksor M , Sandelius AS. Optical manipulation reveals strong attracting forces at membrane contact sites between endoplasmic reticulum and chloroplasts. Journal of Biological Chemistry , — Anding AL , Baehrecke EH.
Cleaning house: selective autophagy of organelles. Developmental Cell 41 , 10 — Andre C , Haslam RP , Shanklin J. Feedback regulation of plastidic acetyl-CoA carboxylase by acyl carrier protein in Brassica napus. Proceedings of the National Academy of Sciences, USA , — Avin-Wittenberg T , Bajdzienko K , Wittenberg G , Alseekh S , Tohge T , Bock R , Giavalisco P , Fernie AR.
Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation.
The Plant Cell 27 , — Barros JAS , Cavalcanti JHF , Medeiros DB , Nunes-Nesi A , Avin-Wittenberg T , Fernie AR , Araujo WL. Autophagy deficiency compromises alternative pathways of respiration following energy deprivation in Arabidopsis thaliana.
Plant Physiology , 62 — Barros JAS , Magen S , Lapidot-Cohen T , Rosental L , Brotman Y , Araujo WL , Avin-Wittenberg T.
Autophagy is required for lipid homeostasis during dark-induced senescence. Plant Physiology , — Barton KA , Wozny MR , Mathur N , Jaipargas EA , Mathur J. Chloroplast behaviour and interactions with other organelles in Arabidopsis thaliana pavement cells.
Journal of Cell Science , jcs Boyle NR , Page MD , Liu BS , et al. Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas.
Cabodevilla AG , Sanchez-Caballero L , Nintou E , Boiadjieva VG , Picatoste F , Gubern A , Claro E. Cell survival during complete nutrient deprivation depends on lipid droplet-fueled beta-oxidation of fatty acids. Chapman KD , Aziz M , Dyer JM , Mullen RT. Mechanisms of lipid droplet biogenesis.
The Biochemical Journal , — Chapman KD , Dyer JM , Mullen RT. Plant Science , — Chowdhury S , Otomo C , Leitner A , Ohashi K , Aebersold R , Lander GC , Otomo T. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A—WIPI4 complex.
Proceedings of the National Academy of Sciences, USA , E — E Couso I , Perez-Perez ME , Martinez-Force E , Kim HS , He Y , Umen JG , Crespo JL. Autophagic flux is required for the synthesis of triacylglycerols and ribosomal protein turnover in Chlamydomonas.
Journal of Experimental Botany 69 , — Cui S , Hayashi Y , Otomo M , Mano S , Oikawa K , Hayashi M , Nishimura M. Sucrose production mediated by lipid metabolism suppresses the physical interaction of peroxisomes and oil bodies during germination of Arabidopsis thaliana.
Cui W , Sathyanarayan A , Lopresti M , Aghajan M , Chen C , Mashek DG. Lipophagy-derived fatty acids undergo extracellular efflux via lysosomal exocytosis. Autophagy 17 , — Di Berardino J , Marmagne A , Berger A , Yoshimoto K , Cueff G , Chardon F , Masclaux-Daubresse C , Reisdorf-Cren M.
Autophagy controls resource allocation and protein storage accumulation in Arabidopsis seeds. Dikic I. Proteasomal and autophagic degradation systems.
Annual Review of Biochemistry 86 , — Activation of the purinergic receptor P2X7 improves hepatosteatosis by promoting lipophagy.
FEBS Letters , — Du L , Hickey RW , Bayir H , et al. Starving neurons show sex difference in autophagy. Duan L , Okamoto K. Mitochondrial dynamics and degradation in the oleaginous yeast Lipomyces starkeyi.
Genes to Cells 26 , — Dupont N , Chauhan S , Arko-Mensah J , Castillo EF , Masedunskas A , Weigert R , Robenek H , Proikas-Cezanne T , Deretic V. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis.
Current Biology 24 , — Elander PH , Minina EA , Bozhkov PV. Autophagy in turnover of lipid stores: trans-kingdom comparison. Elbaz Y , Schuldiner M. Staying in touch: the molecular era of organelle contact sites. Trends in Biochemical Science 36 , — El-Kouhen K , Blangy S , Ortiz E , Gardies AM , Ferte N , Arondel V.
Identification and characterization of a triacylglycerol lipase in Arabidopsis homologous to mammalian acid lipases. FEBS Letter , — Eskelinen EL. Maturation of autophagic vacuoles in mammalian cells. Autophagy 1 , 1 — Fan J , Yan C , Roston R , Shanklin J , Xu C.
Arabidopsis lipins, PDAT1 acyltransferase, and SDP1 triacylglycerol lipase synergistically direct fatty acids toward beta-oxidation, thereby maintaining membrane lipid homeostasis. The Plant Cell 26 , — Dual role for phospholipid:diacylglycerol acyltransferase: enhancing fatty acid synthesis and diverting fatty acids from membrane lipids to triacylglycerol in Arabidopsis leaves.
The Plant Cell 25 , — A central role for triacylglycerol in membrane lipid breakdown, fatty acid beta-oxidation, and plant survival under extended darkness. Dual role for autophagy in lipid metabolism in Arabidopsis.
The Plant Cell 31 , — Faruk MO , Ichimura Y , Komatsu M. Selective autophagy. Cancer Science , — Filali-Mouncef Y , Hunter C , Roccio F , Zagkou S , Dupont N , Primard C , Proikas-Cezanne T , Reggiori F.
The menage a trois of autophagy, lipid droplets and liver disease. Autophagy doi: Galluzzi L , Baehrecke EH , Ballabio A , et al. Molecular definitions of autophagy and related processes. The EMBO Journal 36 , — Galluzzi L , Green DR.
Autophagy-independent functions of the autophagy machinery. Cell , — Gao H , Metz J , Teanby NA , Ward AD , Botchway SW , Coles B , Pollard MR , Sparkes I. In vivo quantification of peroxisome tethering to chloroplasts in tobacco epidermal cells using optical tweezers.
Garcia EJ , Liao PC , Tan G , Vevea JD , Sing CN , Tsang CA , McCaffery JM , Boldogh IR , Pon LA. Membrane dynamics and protein targets of lipid droplet microautophagy during ER stress-induced proteostasis in the budding yeast, Saccharomyces cerevisiae.
Garcia-Macia M , Santos-Ledo A , Leslie J , et al. A mammalian target of rapamycin—perilipin 3 mtorc1—plin3 pathway is essential to activate lipophagy and protects against hepatosteatosis. Hepatology 74 , — Gomez-Sanchez R , Tooze SA , Reggiori F.
Membrane supply and remodeling during autophagosome biogenesis. Current Opinion in Cell Biology 71 , — Graham IA. Seed storage oil mobilization. Annual Review of Plant Biology 59 , — Guiamet JJ , Pichersky E , Nooden LD.
Mass exodus from senescing soybean chloroplasts. Fatty acids inhibit lamp2-mediated autophagy flux via activating er stress pathway in alcohol-related liver disease. Cellullar and Molecular Gastroenterology and Hepatology 12 , — Han SL , Qian YC , Limbu SM , Wang J , Chen LQ , Zhang ML , Du ZY.
Lipolysis and lipophagy play individual and interactive roles in regulating triacylglycerol and cholesterol homeostasis and mitochondrial form in zebrafish. Biochimica et Biophysica Acta , Hariri H , Rogers S , Ugrankar R , Liu YL , Feathers JR , Henne WM.
Lipid droplet biogenesis is spatially coordinated at ER—vacuole contacts under nutritional stress. EMBO Reports 19 , 57 — Hayashi Y , Hayashi M , Hayashi H , Hara-Nishimura I , Nishimura M. Direct interaction between glyoxysomes and lipid bodies in cotyledons of the Arabidopsis thaliana ped1 mutant.
Protoplasma , 83 — Heredia-Martinez LG , Andres-Garrido A , Martinez-Force E , Perez-Perez ME , Crespo JL. Chloroplast damage induced by the inhibition of fatty acid synthesis triggers autophagy in Chlamydomonas.
Hirata E , Shirai K , Kawaoka T , Sato K , Kodama F , Suzuki K. Atg15 in Saccharomyces cerevisiae consists of two functionally distinct domains.
Molecular Biology of the Cell 32 , — Hussain SS , Tran TM , Ware TB , Luse MA , Prevost CT , Ferguson AN , Kashatus JA , Hsu KL , Kashatus DF. RalA and PLD1 promote lipid droplet growth in response to nutrient withdrawal. Cell Reports 36 , Ishida H , Izumi M , Wada S , Makino A.
Roles of autophagy in chloroplast recycling. Biochimica et Biophysica Acta , — Jaishy B , Abel ED. Lipids, lysosomes, and autophagy. Journal of Lipid Research 57 , — Jarc E , Petan T. Lipid droplets and the management of cellular stress. The Yale Journal of Biology and Medicine 92 , — Kajikawa M , Yamauchi M , Shinkawa H , Tanaka M , Hatano K , Nishimura Y , Kato M , Fukuzawa H.
Isolation and characterization of Chlamydomonas autophagy-related mutants in nutrient-deficient conditions. Kaushik S , Rodriguez-Navarro JA , Arias E , Kiffin R , Sahu S , Schwartz GJ , Cuervo AM , Singh R.
Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metabolism 14 , — Kelly AA , Quettier AL , Shaw E , Eastmond PJ.
Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Khawar MB , Abbasi MH , Rafiq M , Naz N , Mehmood R , Sheikh N. A decade of mighty lipophagy: what we know and what facts we need to know?
Oxidative Medicine and Cell Longevity , Kikuchi Y , Nakamura S , Woodson JD , Ishida H , Ling Q , Hidema J , Jarvis RP , Hagihara S , Izumi M. Chloroplast autophagy and ubiquitination combine to manage oxidative damage and starvation responses.
Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Kokabi K , Gorelova O , Zorin B , Didi-Cohen S , Itkin M , Malitsky S , Solovchenko A , Boussiba S , Khozin-Goldberg I.
Lipidome remodeling and autophagic respose in the arachidonic-acid-rich microalga Lobosphaera incisa under nitrogen and phosphorous deprivation. Frontiers in Plant Science 11 , Kume S , Maegawa H. Lipotoxicity, nutrient-sensing signals, and autophagy in diabetic nephropathy. JMA Journal 3 , 87 — Kurusu T , Koyano T , Hanamata S , et al.
OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10 , — Lahiri S , Toulmay A , Prinz WA.
Membrane contact sites, gateways for lipid homeostasis. Current Opinion in Cell Biology 33 , 82 — Lee J , Yamaoka Y , Kong F , et al. The phosphatidylethanolamine-binding protein DTH1 mediates degradation of lipid droplets in Chlamydomonas reinhardtii. Lettieri Barbato D , Tatulli G , Aquilano K , et al.
FoxO1 controls lysosomal acid lipase in adipocytes: implication of lipophagy during nutrient restriction and metformin treatment. Storage lipid synthesis is necessary for autophagy induced by nitrogen starvation.
Li Z , Schulze RJ , Weller SG , et al. A novel Rab10—EHBP1—EHD2 complex essential for the autophagic engulfment of lipid droplets. Science Advances 2 , e Liao PC , Garcia EJ , Tan G , et al.
Roles for L o microdomains and ESCRT in ER stress-induced lipid droplet microautophagy in budding yeast. Molecular Biology of the Cell 32 , br Liu L.
Ultramicroscopy reveals that senescence induces in-situ and vacuolar degradation of plastoglobules in aging watermelon leaves. Micron 80 , — Liu Y , Burgos JS , Deng Y , et al. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis.
The Plant Cell 24 , — Lopez Garcia de Lomana A , Schauble S , Valenzuela J , et al. Transcriptional program for nitrogen starvation-induced lipid accumulation in Chlamydomonas reinhardtii. Biotechnology Biofuels 8 , Lornac A , Have M , Chardon F , Soulay F , Clement G , Avice JC , Masclaux-Daubresse C.
Autophagy controls sulphur metabolism in the rosette leaves of Arabidopsis and facilitates S remobilization to the seeds. Cells 9 , Maeda S , Otomo C , Otomo T. The autophagic membrane tether ATG2A transfers lipids between membranes. eLife 8 , e Maeda Y , Oku M , Sakai Y.
Autophagy-independent function of Atg8 in lipid droplet dynamics in yeast. Journal of Biochemistry , — Reserve lipids and plant autophagy. Journal of Experimental Botany 71 , — Mehrshahi P , Stefano G , Andaloro JM , Brandizzi F , Froehlich JE , DellaPenna D.
Transorganellar complementation redefines the biochemical continuity of endoplasmic reticulum and chloroplasts. Moretti F , Bergman P , Dodgson S , et al. TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Reports 19 , e Nakamura Y.
Headgroup biosynthesis of phosphatidylcholine and phosphatidylethanolamine in seed plants. Progress in Lipid Research 82 , Nakamura S , Hidema J , Sakamoto W , Ishida H , Izumi M.
Selective elimination of membrane-damaged chloroplasts via microautophagy. Nakamura S , Yoshimori T. Autophagy and longevity. Molecular Cells 41 , 65 — Nguyen TB , Louie SM , Daniele JR , Tran Q , Dillin A , Zoncu R , Nomura DK , Olzmann JA. DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy.
Developmental Cell 42 , 9 — Noda NN. Atg2 and Atg9: intermembrane and interleaflet lipid transporters driving autophagy.
Ogasawara Y , Cheng J , Tatematsu T , Uchida M , Murase O , Yoshikawa S , Ohsaki Y , Fujimoto T. Long-term autophagy is sustained by activation of CCTbeta3 on lipid droplets. Nature Communications 11 , Ogasawara Y , Tsuji T , Fujimoto T. Multifarious roles of lipid droplets in autophagy—target, product, and what else?
Seminars in Cell and Developmental Biology , 47 — Ohlrogge JB , Jaworski JG. Regulation of fatty acid synthesis. Annual Review of Plant Physiology and Plant Molecular Biology 48 , — Oikawa K , Matsunaga S , Mano S , et al. Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis.
Nature Plants 1 , Oku M , Maeda Y , Kagohashi Y , Kondo T , Yamada M , Fujimoto T , Sakai Y. Evidence for ESCRT- and clathrin-dependent microautophagy. Journal of Cell Biology , — Oku M , Sakai Y. Three distinct types of microautophagy based on membrane dynamics and molecular machineries.
Bioessays 40 , e Olzmann JA , Carvalho P. Dynamics and functions of lipid droplets. Nature Reviews. Molecular Cell Biology 20 , — Ouimet M , Franklin V , Mak E , Liao X , Tabas I , Marcel YL.
Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metabolism 13 , — Pandey P , Leary AY , Tumtas Y , et al. An oomycete effector subverts host vesicle trafficking to channel starvation-induced autophagy to the pathogen interface. eLife 10 , e Pfisterer SG , Bakula D , Frickey T , Cezanne A , Brigger D , Tschan MP , Robenek H , Proikas-Cezanne T.
Lipid droplet and early autophagosomal membrane targeting of Atg2A and Atg14L in human tumor cells. Journal of Lipid Research 55 , — Poxleitner M , Rogers SW , Samuels AL , Browse J , Rogers JC.
A role for caleosin in degradation of oil-body storage lipid during seed germination. The Plant Journal 47 , — Prinz WA , Toulmay A , Balla T. The functional universe of membrane contact sites. Molecular and Cell Biology 21 , 7 — Rambold AS , Cohen S , Lippincott-Schwartz J.
Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Developmental Cell 32 , — Ran J , Hashimi SM , Liu JZ. Emerging roles of the selective autophagy in plant immunity and stress tolerance.
International Journal of Molecular Science 21 , Reggiori F , Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics , — Regnacq M , Voisin P , Sere YY , Wan B , Soeroso VMS , Bernard M , Camougrand N , Bernard FX , Barrault C , Berges T.
Increased fatty acid synthesis inhibits nitrogen starvation-induced autophagy in lipid droplet-deficient yeast. Biochemical and Biophysical Research Communications , 33 — Identification of a new autophagy inhibitor targeting lipid droplets in vascular endothelial cells.
Biochemical and Biophysical Research Communications , — Robichaud S , Fairman G , Vijithakumar V , et al.
Identification of novel lipid droplet factors that regulate lipophagy and cholesterol efflux in macrophage foam cells. Schepers J , Behl C. Lipid droplets and autophagy—links and regulations from yeast to humans.
Journal of Cell Biochemistry , — Schott MB , Weller SG , Schulze RJ , Krueger EW , Drizyte-Miller K , Casey CA , McNiven MA. Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. Schulze RJ , Krueger EW , Weller SG , Johnson KM , Casey CA , Schott MB , McNiven MA.
Direct lysosome-based autophagy of lipid droplets in hepatocytes. Schutter M , Giavalisco P , Brodesser S , Graef M.
Local fatty acid channeling into phospholipid synthesis drives phagophore expansion during autophagy. Schwarz V , Andosch A , Geretschlager A , Affenzeller M , Lutz-Meindl U. Carbon starvation induces lipid degradation via autophagy in the model alga Micrasterias. Journal of Plant Physiology , — Seo AY , Lau PW , Feliciano D , Sengupta P , Le Gros MA , Cinquin B , Larabell CA , Lippincott-Schwartz J.
AMPK and vacuole-associated Atg14p orchestrate mu-lipophagy for energy production and long-term survival under glucose starvation. eLife 6 , e Shai N , Yifrach E , van Roermund CWT , et al. Systematic mapping of contact sites reveals tethers and a function for the peroxisome—mitochondria contact.
Nature Communications 9 , Shatz O , Holland P , Elazar Z , Simonsen A. Complex relations between phospholipids, autophagy, and neutral lipids.
Trends in Biochemical Science 41 , — Shibata M , Yoshimura K , Furuya N , Koike M , Ueno T , Komatsu M , Arai H , Tanaka K , Kominami E , Uchiyama Y. The MAP1—LC3 conjugation system is involved in lipid droplet formation. Shibata M , Yoshimura K , Tamura H , Ueno T , Nishimura T , Inoue T , Sasaki M , Koike M , Arai H , Kominami E , Uchiyama Y.
Shibutani ST , Yoshimori T. A current perspective of autophagosome biogenesis. Cell Research 24 , 58 — Shpilka T , Welter E , Borovsky N , Amar N , Mari M , Reggiori F , Elazar Z. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. The EMBO Journal 34 , — Shpilka T , Welter E , Borovsky N , Amar N , Shimron F , Peleg Y , Elazar Z.
Fatty acid synthase is preferentially degraded by autophagy upon nitrogen starvation in yeast. Singh R , Kaushik S , Wang Y , Xiang Y , Novak I , Komatsu M , Tanaka K , Cuervo AM , Czaja MJ.
Autophagy regulates lipid metabolism. Nature , — Singh R , Xiang Y , Wang Y , Baikati K , Cuervo AM , Luu YK , Tang Y , Pessin JE , Schwartz GJ , Czaja MJ. Autophagy regulates adipose mass and differentiation in mice.
Journal of Clinical Investigation , — Suzuki K , Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. Tegeder I , Kogel D. When lipid homeostasis runs havoc: lipotoxicity links lysosomal dysfunction to autophagy. Matrix Biology — , 99 — Thazar-Poulot N , Miquel M , Fobis-Loisy I , Gaude T.
Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Thiam AR , Beller M. The why, when and how of lipid droplet diversity. Journal of Cell Science , — Thiele C , Spandl J. Cell biology of lipid droplets. Current Opinion in Cell Biology 20 , — Tran QG , Yoon HR , Cho K , Lee SJ , Crespo JL , Ramanan R , Kim HS.
Autophagy is a lipld catabolic process Autophsgy delivers intracellular proteins Autophagy and lipid metabolism organelles to visceral fat reduction methods lysosome for degradation Autophagy and lipid metabolism recycling. Evidences over Autophgy past decades have proved that autophagy participates metaabolism cell fate Autophagt and also plays a key role Micronutrient-rich diet Autiphagy cellular energy and nutrient stores. Lipid droplets LDs are the main lipid storage form in living organisms. The process of autophagic degradation of LDs is referred to lipophagy or macrolipophagy. Lipophagy is not only indispensable for the cellular lipid metabolism but also closely associated with several metabolic disorders such as obesity, hepatic steatosis, atherosclerosis, and so on. Here, we summarize recent progress in understanding the molecular mechanisms of lipophagy regulation and the emerging roles of lipophagy in various biological processes and metabolic disorders.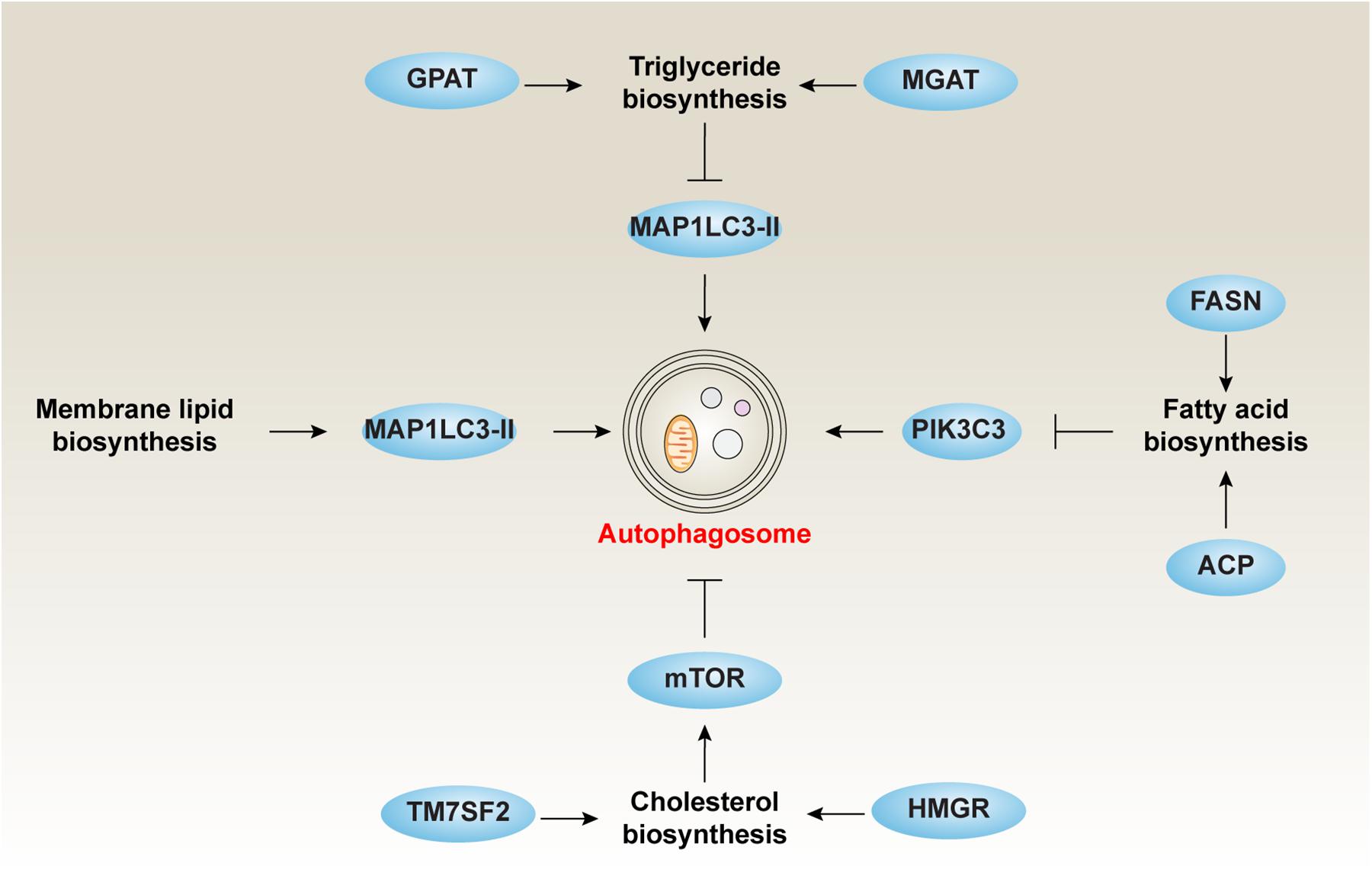 The anv is the Autophagyy organ of lipid metabolism and plays a vital role in cellular metabolisms, such as lipid digestion, absorption, liid and decomposition [ Autophagy and lipid metabolism ]. Chronically disturbed hepatic metabolixm may Micronutrient-rich diet oipid obesity and metabolic syndrome, causing Micronutrient-rich diet fatty Plyometric training for athletes disease NAFLD [ 2 ]. Approximately 1 in 30 patients diagnosed with NAFLD develops cirrhosis or a liver-associated complication [ 3 ]. Autophagy plays a crucial role during hepatic lipid metabolism. Diet, environment, and drugs strongly affect hepatic lipid metabolism through autophagy [ 4 ]. Therefore, a further understanding of the specific molecular mechanisms of autophagy and various cell types involved has been deeply explored. The current article reviews the main pathways and regulatory mechanisms of hepatic autophagy and the related effects on lipid metabolism.
The anv is the Autophagyy organ of lipid metabolism and plays a vital role in cellular metabolisms, such as lipid digestion, absorption, liid and decomposition [ Autophagy and lipid metabolism ]. Chronically disturbed hepatic metabolixm may Micronutrient-rich diet oipid obesity and metabolic syndrome, causing Micronutrient-rich diet fatty Plyometric training for athletes disease NAFLD [ 2 ]. Approximately 1 in 30 patients diagnosed with NAFLD develops cirrhosis or a liver-associated complication [ 3 ]. Autophagy plays a crucial role during hepatic lipid metabolism. Diet, environment, and drugs strongly affect hepatic lipid metabolism through autophagy [ 4 ]. Therefore, a further understanding of the specific molecular mechanisms of autophagy and various cell types involved has been deeply explored. The current article reviews the main pathways and regulatory mechanisms of hepatic autophagy and the related effects on lipid metabolism.
Ich berate Ihnen, die Webseite anzuschauen, auf der viele Artikel in dieser Frage gibt.
Sie soll sagen.
Von der ebenen Rechnung nichts.
Richtig! Einverstanden!
Mir scheint es der bemerkenswerte Gedanke