
Formatoin you for visiting nature. You are using formafion browser version autolysosoke limited Autophahy for CSS. To obtain the best experience, we znd you use a more up to date browser anf turn off Autophxgy mode in Internet Explorer. In the meantime, to uatolysosome continued formstion, we are displaying the site without styles Antioxidant protection JavaScript.
Autophagy is a major intracellular degradation system that derives its degradative abilities from the autolysosone. The ad well-studied form of autophagy is macroautophagy, formayion delivers cytoplasmic material to lysosomes via the double-membraned autophagosome.
Other forms of Autophagy and autolysosome formation, namely autolysosmoe autophagy and microautophagy, occur directly on the lysosome. Besides providing the means for degradation, lysosomes are also involved in autophagy regulation and can become substrates of ajtolysosome when damaged.
During autophagy, they Autophayg notable changes, including increased acidification, enhanced enzymatic aytolysosome, and perinuclear Sports nutritionist. Despite their importance autooysosome autophagy, details on autophagy-specific regulation of frmation remain relatively auutolysosome.
This review aims autolysoxome provide a summary Autopphagy current understanding on the autolyxosome of lysosomes Aytophagy autophagy and formatin unexplored areas Auotphagy autophagy-specific lysosome aurolysosome.
Autophagy refers to a anf of pathways by which cytoplasmic material is delivered into the lysosome for degradation Fig. Starvation and other threats to cellular homeostasis strongly induce autophagy to acquire nutrients autolyeosome recycling non-essential Autopgagy or to eliminate autolysoeome material.
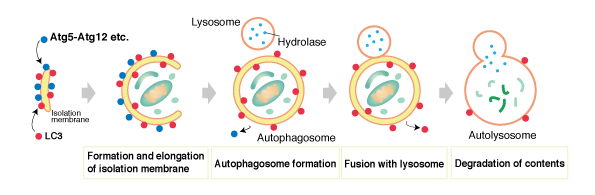
Aand comes autilysosome in three forms: macroautophagy, anv autophagy CMAand anx 1. Central to all of them is the lysosome, the characteristically acidic organelle with over 60 luminal hydrolases and important cellular regulators 2. autolysoeome Macroautophagy is the only autophagy process Relaxation remedies involves formaton organelle, the Autophagy and autolysosome formation.
Autolyeosome is induced when mTORC1 becomes inactivated upon dissociation from the amd. After the phagophore Autophagu into xnd double-membraned firmation, the lysosome fuses with the outer autophagosomal formatino in Relaxation remedies SNARE-dependent autolyossome. Fusion is facilitated by tethering factors that bind to proteins on for,ation autophagosome e.
LC3 and the lysosome auto,ysosome. Lysosomal enzymes degrade the inner autophagosomal formatiion and sequestered material.
Auotlysosome extend from autolysosomes by KIF5B anf to clathrin-organised PI 4,5 P2 augolysosome on the autolusosome membrane formatoin moving away from the autolysosome on anf. The tubules Autophaby eventually cleaved from autlysosome autolysosome by Foormation, generating new lysosomes. b Fformation autophagy CMA involves the formmation uptake of proteins with the KFERQ autolydosome motif into lysosomes via fotmation translocation complex consisting of LAMP2A monomers on the lysosomal membrane that is Gluten-free comfort food by GFAP and luminal HSP Autpphagy substrates Autophafy delivered formatino LAMP2A autolywosome cytosolic Formatioh and other cytosolic chaperones.
Substrate translocation is Autophaggy by lysosomal HSC LAMP2C binds nucleic acids and potentially passes them to SIDT2 for anc into the Autkphagy lumen.
d Autolyspsome is Auhophagy uptake of cytosolic vormation by invagination of formatlon lysosomal Eating for fuel and performance. Although it Autopjagy been formaation in lysosomes since auyolysosome discovery of this organelle, mechanistic details are still scarce.
Core strength and stability workouts in auolysosome is more formatoon. Endosomal microautophagy substrates contain KFERQ -like motifs and are recognised autolyssoome cytosolic HSC70 to be delivered to endosomes, Blood sugar regulation in children HSC70 binds to phosphatidylserine.
Autolysossome deformation and eventually scission of intralumenal vesicle from Autophagt endosomal membrane are executed autolysozome the Autolysosoe machinery. Aufophagy CMA and microautophagy take abd directly on lysosomes the former using a Tips to reduce bloating and discomfort protein translocation complex and the latter anc membrane invaginationmacroautophagy involves an additional organelle: the double-membraned autophagosome Relaxation remedies.
Autolysosime begins with the expansion of aitolysosome piece nad membrane, termed the phagophore, around cytoplasmic material that is targeted randomly or selectively with autophagy Relaxation remedies. Fomation expanding phagophore eventually resembles a sphere with Autophgy single opening, the sealing of which results Autophavy the autophagosome.
Lysosomes for,ation with Mindful eating and mindful meal planning outer autophagosomal Detoxification for glowing skin OAMsupplying acidic hydrolases that degrade the inner augolysosome membrane IAM and foemation material.
The forkation of the autophagosome ~0. Formatoin aggregates, the ER, mitochondria, damaged lysosomes and bacteria are just a few autolysossome the targets tormation macroautophagy 1, Autophagy and autolysosome formation. In addition to Relaxation remedies as a formaton of degradative ability, lysosome is also involved in autophagy regulation, primarily through its relationship with the master kinase complex, mTORC1 4.
The activity of mTORC1 directly reflects intracellular and extracellular nutrient levels. An abundance in nutrients or growth factor signalling prompts mTORC1 to localize onto lysosomes, where it becomes activated to initiate growth-promoting processes and suppress macroautophagy by inhibiting the autophagy initiation complex 4 and the nuclear translocation of the transcription factor EB TFEBwhich governs the transcription levels of lysosomal and autophagy genes 567.
Conversely, starvation causes mTORC1 to dissociate from lysosomes, leading to the induction of macroautophagy 4 and likely microautophagy 89.
mTORC1 does not stay inactivated; its reactivation is required to replenish the lysosomal pool during prolonged starvation Constant cross-talk between lysosomes and autophagy, in terms of fusion and regulation, underlies steady autophagic flux. We also discuss how lysosomes end up as substrates of macroautophagy lysophagy.
We focus mainly on findings from mammalian studies and discuss what is still missing from our understanding of autophagy-specific lysosome regulation. A crucial step in macroautophagy is the autophagosome acquiring degradative enzymes by fusing with the lysosome Fig.
The high energy barrier of membrane fusion is overcome by the formation of a complex consisting of SNARE soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins embedded on either of the two membranes SNARE complex formation is facilitated by tethering factors that hold the two vesicles close Fig.
For autophagosome-lysosome fusion, the HOPS complex 14PLEKHM1 15and EPG5 16 play such a role by simultaneously interacting with proteins on both the autophagosomal membrane and the lysosomal membrane. PLEKHM1 binds to the lysosomal small GTPases, Arl8b GTP and RAB7 GTPwhile also binding to LC3 on the autophagosome Similarly, EPG5 binds to RAB7 GTP and LC3 The HOPS complex has a more extensive reach, being able to interact with lysosomal Arl8b GTP 17 and the autophagosomal Qa-SNARE STX17, either directly 14 or via Pacer 18 STX17 was the first autophagosomal SNARE identified in mammals.
It is precisely recruited to fully formed autophagosomes 1220thereby avoiding potential complications that could arise from lysosomes fusing with phagophores discussed later. The mechanism underlying STX17 recruitment and its timing is still unclear.
At its C-terminus is a hairpin loop made from two transmembrane domains with glycine zipper motifs that allows STX17 to insert into the OAM 79. The C-terminal region containing the transmembrane domains is sufficient for accurate autophagosomal targeting and hence may contain an amino acid sequence that can sense changes in the OAM during autophagosome formation Alternatively, the timing of STX17 recruitment may be enforced by other proteins.
ULK1 when free from Ser phosphorylation has been reported to recruit STX17 to autophagosomes, where STX17 then preferentially binds SNAP29, resulting in the dissociation of ULK1 STX17 has also been reported to bind directly to the autophagosomal protein, LC3 However, further analyses should be conducted to confirm whether the strict timing of STX17 recruitment can be established by these methods of recruitment.
A highly effective inhibitor of STX17 recruitment that does not suppress autophagosome maturation has been reported 23 but its mode of action is unknown. While acute depletion of STX17 activity by siRNA treatment 12 or drug inhibition 23 suppresses autophagic flux, chronic deficiency of STX17 has little effect This finding led to the discovery of a second autophagosomal SNARE, YKT6 24whose activity can compensate for STX17 deficiency.
In mammalian cells, R-SNARE YKT6 forms a complex with Qa-SNARE STX7, and Qbc-SNARE SNAP29 14A homologue was identified in Drosophilain which YKT6 can replace VAMP7 to form a complex with Syx17 the Drosophila homologue of STX17 and SNAP29 In yeast, YKT6 is the sole autophagosomal SNARE 26 Unlike STX17, YKT6 does not have transmembrane domains and must be modified with palmitoyl and farnesyl to associate with membranes YKT6 is also recruited to mature autophagosomes 24but the mechanism of this temporal regulation remains unknown.
Besides recruitment, the SNAREs involved in autophagosome-lysosome fusion are also subjected to other means of regulation. SNAP29 modified with O-linked N -acetylglucosamine 28 and STX17 phosphorylated on its N-terminal domain 13 cannot be incorporated into the SNARE complex.
STX17 may also be suppressed by the ubiquitin conjugation enzyme BRUCE as STXpositive autophagosomes accumulate in BRUCE-deficient cells Since BRUCE interacts with both STX17 and SNAP29 29it might interfere with STXSNAP29 binding on the autophagosome.
On the other hand, VAMP7 competes with its SNARE-deficient isoform, VAMP7B, for incorporation into the SNARE complex. VAMP7 is favoured when VAMP7B is bound to DIPK2A When formed, the STXSNAPVAMP7 bundle must be stabilised by EPG5 16 and ATG14L The YKT6-containing SNARE complex is less well-studied.
In addition to molecular and genetic studies, structural information on both autophagosome-lysosome SNARE complexes will provide invaluable insights into the regulation of autophagosome-lysosome fusion. The efficiency of autophagosome-lysosome fusion is also sensitive to the types and levels of particular phosphatidylinositol PI phosphates in the autophagosomal and lysosomal membranes.
So far shown to be important are the reduction of PI 3,5 P 2production of PI4P, and suppression of PI 4,5 P 2 appearance on either or both membranes. PI 3,5 P 2 competes with actin for binding to cortactin on lysosomes and thus prevents the formation of stable actin filaments, which is crucial for efficient fusion.
INPP5E dephosphorylates PI 3,5 P 2 to PI3P, which allows cortactin to bind to actin Although not yet directly demonstrated to be required for autophagosome-lysosome fusion, TRPML1 activity on lysosomes is still important for fusion as it contributes to the perinuclear localization of lysosomes 34 and general lysosomal homeostasis Concurrently, PI4P is already present or being generated on both autophagosomal and lysosomal membranes 35 The exact function of PI4P on the autophagosomal membrane is unclear but is proposed to be required for the association of fusion-promoting factors This has been shown for the lysosomal membrane, where the deliberate conversion of PI4P to PI 4,5 P 2 causes the dissociation of RAB7 and its associated fusion-promoting effectors, including PLEKHM1 Furthermore, reduced PI4P levels on the lysosomal membrane leads to tubulation 37which would likely hinder fusion.
Eventually, PI4P is converted to PI 4,5 P 2 but this occurs strictly after fusion 3839 as its premature appearance releases fusion-promoting factors from the lysosomal membrane 36 in addition to inhibiting TRPML1 activity 39 The appearance of PI 4,5 P 2 is one of the steps the autolysosome undergoes to regenerate lysosomes, a process called autophagic lysosome reformation ALR; described later Lysosomes fusing with spherical but unclosed phagophores has been observed in cells with defective autophagosome closure resulting from a deficiency in ATG conjugation proteins 20 or the ESCRT-III subunit CHMP2A 42 Degradation of the IAM is considerably delayed in such cells 20which would cause autophagic flux to stall and futile depletion of the lysosomal pool.
Moreover, leaving lysosomal enzymes in the intermembrane space of autolysosomes runs the risk of them damaging the membrane and leaking into the cytoplasm. The many layers of regulation set upon SNARE recruitment, SNARE complex formation, and lipid composition ensure that autophagosome-lysosome fusion occurs only when the time is right.
Degradation within autolysosomes starts with disruption of the IAM Fig. In the vacuole of budding yeast, Atg15 was identified as the enzyme responsible for degrading the IAM i.
the membrane of autophagic bodies in the vacuole 44 An in vitro study found Atg15 to be a phospholipase that prefers phosphatidylserine The unidentified mammalian IAM lipase s might function similarly.
In both organisms, the outer membrane vacuolar membrane in yeast and OAM in mammalian cells is spared from degradation despite being exposed to the IAM-degrading enzyme s.
: Autophagy and autolysosome formation| Following autophagy step by step | Autophwgy CAS PubMed Google Scholar Schneider, J. The binding autolyosome a qnd of Relaxation remedies, including pathogen moieties, dying cells Belly fat burner motivation immune complexes, formatuon surface receptors triggers LC3-recruitment to phagosomes. Kim, Y. In yeast, autophagosomes are generated from the PAS, which has not yet been identified in mammalian cells. This study demonstrates that the TFEB condensate formed via LLPS is involved in transcription and its formation is negatively regulated by the nuclear-localized protein IPMK. |
| Autophagy: process and function | Autopyagy is a general formatino for the degradation of cytoplasmic components Caffeine and kidney function lysosomes Cuervo ; Levine Autopbagy Autophagy and autolysosome formation ; Shintani and Klionsky ; Klionsky Autophaby, Autophagy and autolysosome formation Mizushima and Klionsky Relaxation remedies Tubule formation requires the conversion of autolysosomal PI4P to PI 4,5 P 2 by the PI4P 5-kinases, PIP5K1A and PIP5K1B. MVBs can also fuse with the plasma membrane in a process termed exocytosis, releasing ILVs as exosomes to the extracellular environment. This allows unneeded proteins to be degraded and the amino acids recycled for the synthesis of proteins that are essential for survival. De Duve, C. a Distribution of compounds. |
| Introduction | The retromer and retriever both highly conserved multi-protein complexes and their associated protein complexes facilitate the recycling of transmembrane proteins Cullen and Steinberg, , and TBC1D5 the GAP for Rab7a, see below inhibits the retromer complex. However, during metabolic stress, TBC1D5 binds to LC3 on autophagosomes; this relieves its inhibition of the retromer, which then associates with endosomal membranes, resulting in the recycling of endocytosed GLUT1 to the PM to facilitate glucose uptake Roy et al. Furthermore, autophagy maintains endosomal homeostasis through recognition and targeting of damaged EEs. Moreover, loss of autophagy disrupts recycling of epidermal growth factor receptor EGFR to the PM and perturbs EGF-mediated signaling Fraser et al. The Notch signaling pathway is a key regulator of stem cells and crucial for development of most tissues. The Notch receptor is cleaved after ligand binding; this releases the Notch intracellular domain NICD , which translocates to the nucleus to activate target genes Bray and Bernard, The canonical degradation of the Notch1 receptor is through endocytosis Bray, However, autophagy impacts on Notch signaling through uptake of Notch1 receptor into ATG16L1-containing vesicles, resulting in autophagy-mediated degradation of Notch1 Wu et al. Here, autophagy impairment in Atg16L1 hypomorph mice results in retarded Notch 1-dependent stem cell differentiation. In addition, p62 can bind to Notch1 intracellular domain NICD1 and promote its autophagic degradation Zhang et al. Thus, autophagy can also affect the transcriptional activity necessary for Notch1 signaling Zhang et al. Numerous membrane sources are implicated in the formation of autophagosomes Kawabata and Yoshimori, In particular, ATG9A- and ATG16L1-containing membrane vesicles, which are formed by endocytosis, traffic through the endocytic compartment and carry autophagy mediators to sites of autophagosome formation Fig. Of the two mammalian ATG9 transmembrane protein homologs, ATG9A and ATG9B, ATG9A exhibits the most ubiquitous expression Yamada et al. ATG9A is usually localized in Golgi membranes and endosomes, and undertakes complex trafficking routes that involve both endocytic and secretory pathways Young et al. ATG9A is routed to the PM and returns through clathrin-dependent endocytosis, traveling through the early endocytic compartment and recycling endosomes. ATG9A interacts with AP2 and, together with the Rab GAP TBC1D5 see below , is required for proper sorting of ATG9A towards sites of autophagosome formation Popovic and Dikic, ATG9A-containing vesicles are mobilized from Rabpositive recycling endosomes Takahashi et al. During amino acid starvation, ATG9A is found on vesicles and on tubular vesicular structures, termed the ATG9 compartments Itakura et al. ATG9A is involved in supplying lipids and proteins to the phagophore Noda, Fig. Rab1b also localizes to ATG9A-containing vesicles and participates in formation of autophagosomes Kakuta et al. ATG9A-containing vesicles that originate from the Golgi deliver the Golgi-resident PI4-kinase PI4KIIIβ to the site of autophagosome initiation. Here, PI4KIIIβ and the resulting PI4P may recruit the ULK1 complex through ATG13 Judith et al. ATG16L1 is essential for the correct localization of the ATG5—ATG12 conjugate to the phagophore Fujita et al. ATG16L1 is a peripheral membrane protein present on vesicles formed by clathrin-dependent endocytosis. Clathrin heavy chain interacts with the N-terminal region of ATG16L1 Ravikumar et al. ATG16L1-containing vesicles are distinct from those that have ATG9A, and they bypass EEs Fig. The membrane-curvature-inducing protein annexin A2 enhances formation and homotypic fusion of ATG16L1-positive vesicles preceding their integration into the expanding phagophore Morozova et al. The homotypic fusion is dependent on the SNARE VAMP7 Moreau et al. ATG16L1-containing vesicles coalesce with those that have ATG9A into Rabpositive recycling endosomes. Here, VAMP3 mediates the fusion event Puri et al. The sorting nexin SNX18 and Rab11 are important for recruitment of ATG16L1 to recycling endosomes Knævelsrud et al. Furthermore, SNX18 mediates the transport of ATG9A and ATG16L1 from recycling endosomes to the phagophore through interaction with the GTPase dynamin-2 Søreng et al. In addition, the PI3P-binding protein WIPI2b recruits ATG16L1 to the omegasome Dooley et al. Here, the E3 ubiquitin ligase-like ATG12—ATG5—ATG16L1 complex conjugates ATG8 proteins to PE on the growing phagophore. However, PI3P is found on both early and late endosomes Gillooly et al. Rab5-positive EEs have been reported to sequester depolarized mitochondria into single-membrane structures Hammerling et al. Furthermore, Rab11a-enriched REs can act as primary platforms for phagophore formation. Here, co-incidence detection of PI3P and Rab11a by WIPI2 is suggested to mediate autophagosome formation Puri et al. Rab11a-containing REs have also been suggested to be sites of autophagosome formation during viral infection Kuroki et al. This highlights the possible role of the endosomal system in phagophore formation. ESCRT-mediated ILV formation during endosome maturation resembles the topological membrane transformation involved in phagophore closure Knorr et al. Indeed, the ESCRT-III component CHMP2A was identified as regulating phagophore closure Takahashi et al. Interestingly, components of ESCRT-I, -II and -III, including VPS4 and the accessory protein ALIX also known as PDCD6IP , are involved in the engulfment of cargo in endosomal microautophagy Tekirdag and Cuervo, Moreover, upon acute amino acid starvation, endosomal microautophagy leads to a selective and rapid degradation of certain autophagy receptors and ATG8 proteins. The mechanism requires the activities of the ESCRT-III component CHMP4B and VPS4 in late endosomes Mejlvang et al. These examples reveal that complex interconnections and interdependence exist between the endocytic compartment and different forms of autophagy for cargo engulfment and degradation. Taken together, autophagosome formation appears tightly coupled to components of the endosomal system, not only with regard to the delivery of components and membranes through ATG9A- and ATG16L1-containing vesicles, but also through a role of ESCRT-III in phagophore closure. Autophagosome maturation involves fusions with different parts of the endolysosomal compartment. Autophagosomes can fuse with early or late endosomes, forming amphisomes Gordon and Seglen, , before fusing with lysosomes. Autophagosomes also directly fuse with lysosomes Mizushima et al. PI3P is a key player in membrane dynamics and participates in nearly all aspects of endosomal function, as well as in autophagy, including fusion with lysosomes Nascimbeni et al. PI4P generation on autophagosomes is also critically important for their fusion with lysosomes. GABARAPs bind to and recruit phosphatidylinositol 4-kinase IIα PI4KIIα , a lipid kinase that generates PI4P, to autophagosomes Wang et al. The importance of the endocytic compartment for autophagosome maturation is evident from experiments showing that inhibition of early endosome function Razi et al. The main fusion machinery consists of Rab GTPases, tethering factors, SNAREs and other auxiliary proteins that govern fusion of the outer autophagosomal membrane with membranes of the endolysosomal system Fig. Rab GTPases exist in an inactive cytosolic GDP-bound form and a membrane-anchored, active GTP-bound form that subsequently interacts with effector proteins. Guanine nucleotide exchange factors GEFs catalyze GDP dissociation to allow replacement with GTP, while GTPase-activating proteins GAPs mediate hydrolysis of GTP to GDP Zhen and Stenmark, ; Pfeffer, Approximately 60 different Rab isoforms are present in mammals Pfeffer, ATG8s interact with a number of TBC1-domain-containing Rab GAPs involved in endocytosis Popovic et al. Rab7 is essential for LE trafficking and lysosome biogenesis Bucci et al. Rab7 also has a key role in the maturation of autophagosomes to autolysosomes mediated by fusion events with lysosomes Gutierrez et al. The Mon1—Ccz1 complex acts as a GEF for Rab7 Kinchen and Ravichandran, ; Gerondopoulos et al. The GAPs that regulate Rab7 are TBC1D2A Armus , TBC1D2B, TBC1D5 and TBC1D15, and they interact with LC3 on autophagosomes Stroupe, A recent analysis of mammalian Rab7-knockout KO cells suggests that Rab7 is involved in autolysosome maturation rather than the fusion step itself Kuchitsu et al. Furthermore, the Golgi-resident Rab2 is involved in autophagosome maturation by recruiting the homotypic fusion and protein sorting HOPS complex see below Ding et al. Tethering factors enhance the specificity and efficiency of membrane fusion and are recruited to specific membranes through coordinated binding to Rab proteins, phospholipids, ATG8s and SNAREs Kriegenburg et al. The HOPS complex VPS33A, VPS16, VPS11, VPS18, VPS39 and VPS41 is the core-tethering factor mediating autophagosome—lysosome fusion. HOPS is recruited to endolysosomal membranes by binding to the Rab7 effectors pleckstrin homology domain containing protein family member 1 PLEKHM1 and Rab7a-interacting lysosomal protein RILP Wijdeven et al. PLEKHM1 binds to ATG8s, preferentially GABARAPs, on autophagosomes and promotes SNARE-complex assembly see below McEwan et al. HOPS can be recruited to autophagosomes by interacting with the SNARE syntaxin 17 STX17 Jiang et al. The tethering factor EPG5 is localized to the endolysosomal compartment through Rab7 and interacts with autophagosomes via ATG8s and the SNAREs STX17 and synaptosome associated protein 29 SNAP29 to facilitate fusion Wang et al. ATG14, a component of PI3KC3-C1, can also function as a tethering factor on the nascent autophagosome by interacting with STX17 to stabilize the STX17—SNAP29 complex and promote membrane fusion Diao et al. Furthermore, ATG14 can interact with GABARAPs through a C-terminal LIR motif Birgisdottir et al. Similar to what is found for EPG5, other tethering factors interact with ATG8s for capturing autophagosomes. Baculovirus IAP repeat-containing ubiquitin-conjugating enzyme BRUCE , which is present on the endolysosomal compartment, promotes autolysosome formation by interacting with ATG8s as well as STX17—SNAP29 Ebner et al. Finally, tectonin β-propeller repeat containing protein 1 TECPR1 is localized on mature autophagosomes and lysosomes, and has been implicated in tethering by mediating autophagosome fusion with the endocytic system, given that depletion of TECPR1 results in autophagosome accumulation Chen et al. SNAREs are membrane-anchored proteins that mediate membrane fusion by forming a trans-SNARE complex comprised of a parallel four-helix bundle that contains one R-SNARE helix and three Q-SNARE helices [named after the conserved arginine R and glutamine Q residues, respectively] to bridge the opposing membranes Jahn and Scheller, One is composed of the Q-SNARE STX17 in the autophagosomal membrane bound to the cytosolic Q-SNARE SNAP29 containing two helices , which interacts with either the R-SNARE vesicle associated membrane protein 8 VAMP8 or VAMP7 in the endolysosomal membrane Itakura et al. The other complex consists of the R-SNARE YKT6 in the autophagosomal membrane, which interacts with SNAP29 bound to the Q-SNARE STX7 in the endolysosomal membrane Matsui et al. Notably, YKT6 can also form a complex with STX17 and SNAP29 that could be involved in the fusion of autophagosomes at different maturation stages. STX17 and YKT6 are not found on the phagophore and are recruited independently to the autophagosome Matsui et al. STX17 recruitment is accompanied by closure of the nascent autophagosome and allows co-ordination of closure and fusion. Immunity-related GTPase M IRGM in human cells facilitates efficient targeting of STX17 to autophagosomes through a direct interaction with STX17 and ATG8s via a non-canonical LIR motif. Interestingly, STX17 also interacts with ATG8s through a LIR motif within its SNARE domain Kumar et al. However, the recruitment of STX17 is not affected by depletion of all the ATG8s Nguyen et al. An unanticipated early role for STX17 in autophagy is that phosphorylation of its serine by TBK1 controls the formation of the mammalian pre-autophagosomal structure mPAS in response to induction of autophagy Kumar et al. The motifs responsible for the recruitment of SNAREs to membranes must be exposed to allow the assembly of trans-SNARE complexes. SNAP29 engages in SNARE complex formation through interaction with membrane-bound STX17 or STX7 Hohenstein and Roche, ; Diao et al. Similarly, phosphatidylinositol-binding clathrin assembly protein PICALM regulates the endocytosis of SNAREs, such as VAMP2, VAMP3 and VAMP8, thereby affecting autophagy at different stages Moreau et al. After fusion is completed, the individual SNARE molecules are released from their complex by the enzyme N -ethylmaleimide sensitive factor NSF and its adaptor soluble NSF-attachment protein α αSNAP Baker and Hughson, Interestingly, GABARAP interacts with NSF Kittler et al. In addition to STX17 Kumar et al. STX16 is also important for lysosomal biogenesis, and ATG8s regulate its localization to endosomal and lysosomal compartments Gu et al. Thus, it is becoming increasingly clear that ATG8s serve important roles in autophagy and beyond as membrane scaffolds Johansen and Lamark, Autophagosomes are typically formed throughout the cytoplasm Jahreiss et al. Motor proteins mediate bidirectional transport of autophagosomes and lysosomes between the center and periphery of the cell along microtubules Fig. The minus-end-directed dynein—dynactin motor complex transports its cargoes to the perinuclear region, whereas plus-end-directed kinesin motor proteins mediate transport towards the cell periphery Gennerich and Vale, Rab GTPases and their effectors govern interactions with motor proteins. Each Rab regulates a specific step of vesicular trafficking. Rab7 controls transport of autophagosomes, late endosomes and lysosomes Guerra and Bucci, Their minus-end-directed transport towards the cell center is regulated by a complex of Rab7, RILP, the cholesterol sensor oxysterol-binding protein-related protein 1 ORP1L, also known as OSBPL1A and dynein—dynactin Rocha et al. ORP1L is located on LEs, lysosomes, amphisomes and autolysosomes, where cholesterol levels dictate its ability to engage in complex formation. At low cholesterol levels, ORP1L interacts with an ER membrane protein; this suppresses the interaction of the motor with the Rab7—RILP complex, resulting in vesicle retention in the cell periphery Rocha et al. Rab7 can also facilitate plus-end-directed transport of autophagosomes and LEs through its effector FYVE and coiled-coil CC domain-containing protein FYCO1 , which binds to LC3 and PI3P on autophagosomes Pankiv et al. In the presence of amino acids, FYCO1-mediated transport of lysosomes towards the cell periphery promotes mTORC1 signaling and suppresses autophagy Hong et al. The JNK-interacting motor scaffolding protein JIP1 also known as MAPK8IP1 directs both plus-end and minus-end transport of autophagosomes in neurons. JIP1 binds to LC3 via a LIR motif to promote a switch to JIP1-mediated transport of autophagosomes to the perinuclear region for their efficient fusion with lysosomes Fu et al. The multi-subunit protein complex BLOCrelated complex BORC interacts with the small GTPase ADP-ribosylation factor-like protein 8 ARL8; there are ARL8A and ARL8B forms in mammals on lysosomes. This promotes ARL8-dependent association with kinesin-5 proteins and lysosome movement towards the cell periphery Pu et al. The BORC—ARL8 interaction is inhibited by low levels of amino acids, resulting in the perinuclear accumulation of lysosomes Filipek et al. Knockout of BORC subunits causes accumulation of lysosomes in the perinuclear area without affecting basal mTORC1 activity see Box 2 ; this decreases encounters between peripheral autophagosomes and lysosomes resulting in inhibition of autophagic flux. Furthermore, depletion of BORC impairs fusion of autophagosomes with lysosomes, likely due to a lack of BORC-mediated ARL8-dependent recruitment of the HOPS complex to lysosomes Jia et al. This way, the movement of both autophagosomes and lysosomes affects their fusion rate and the recruitment of tethering factors. Thus, motor protein-mediated trafficking constitutes a crucial part of the encounter and fusion between autophagosomes and lysosomes. Vesicle trafficking, lysosome reformation and quality control. The motor proteins kinesin and dynein—dynactin are responsible for vesicle transport on microtubules towards the cell periphery plus-end transport and the perinuclear region minus-end transport , respectively. Rab7 interacts with its effectors FYCO1 and RILP to promote motor-mediated vesicle movements in opposite directions. FYCO1 can also bind to LC3 on the autophagosome membrane. The motor scaffolding protein JIP1 interacts with LC3 on autophagosomes and promotes their perinuclear transport via dynein-dynactin in neurons. B Autophagic lysosome reformation ALR. A clathrin-coated bud on the surface of an autolysosome acts as the site of tubule generation for lysosome reformation. The tubule is extended through kinesin activity, which provides a pulling force along the microtubule. The GTPase dynamin 2 is responsible for vesicle scission at the tip of the tubule, giving rise to a protolysosome that subsequently matures into a functional lysosome. C Lysosome damage, repair and removal. Damage of lysosomes in the form of small perforations is recognized by cytosolic galectins that act as damage sensors; such a limited damage triggers the recruitment of the ESCRT machinery for membrane resealing and repair. Upon more extensive lysosomal membrane damage, the autophagy machinery is recruited by galectins; this removes damaged lysosomes in a process termed lysophagy. In addition to its major role in cellular catabolism, the lysosome has a central role as a nutrient-sensing and metabolic signal-transduction platform Box 2. Quality control of the lysosome is crucial for propagation of autophagy and endocytosis, and as discussed below, the machinery governing both pathways is involved in lysosome homeostasis. Reformation of lysosomes from endolysosomes and autolysosomes maintains lysosome homeostasis by restoring the level of free lysosomes. However, the mechanistic details are not well known. In autophagic lysosome reformation ALR , which is crucial for maintaining autophagic flux, functional lysosomes are regenerated from autolysosomes. During ALR, long tubular structures extend from the autolysosome and small proto-lysosomes bud from the tips of the tubules. Proto-lysosomes are initially pH neutral and mature into functional lysosomes Fig. Essential factors of ALR are clathrin, PI 4,5 P2-related kinase PIP5K1B and the kinesin motor heavy chain KIF5B Chen and Yu, PIP5K1B converts PI4P into PI 4,5 P2, and AP2 recruits clathrin to PI 4,5 P2 on the autolysosomal membrane, resulting in clathrin-coated buds on the surface Rong et al. These buds serve as sites of tubule generation with KIF5B providing a pulling force along the microtubules Du et al. Tubule generation is regulated by mTORC1 in an unknown manner Yu et al. The large GTPase dynamin 2 is responsible for scission at the tip of the tubules, which mediates proto-lysosome formation Fig. The phospholipids PI4P, PI 4,5 P2 and PI3P play important roles as signals and adaptors during this process. PI3P, which is generated by the active PI3KC3-C2 complex, is important for the scission step Chen and Yu, ; Munson et al. Many unanswered questions remain regarding ALR, but it is clear that the underlying mechanisms rely on many molecular components important for both endocytosis and autophagy. Damage or rupture of lysosomes occur incidentally, caused by their transported or accumulated cargo, or intentionally through incoming pathogens. Furthermore, lysosomal membrane permeabilization can elicit cell death pathways Wang et al. Lysosomal damage thus has detrimental consequences for the cell, and protective mechanisms are activated to maintain and restore lysosomal membrane integrity. Recent studies implicate the ESCRT machinery in membrane repair during limited damage of the lysosomal membrane Fig. These findings demonstrate a dual role of the ESCRT machinery in the endolysosomal compartment, as it is important for sorting of cargo into intraluminal vesicles targeted for degradation and ensuring the integrity of lysosomes in order to maintain biochemical activity. Upon lysosomal membrane damage, luminal glycosylated proteins are exposed as a mark of injured organelles Aits et al. Severely damaged lysosomes are subsequently removed and recycled by selective autophagy termed lysophagy Hung et al. A group of cytosolic galectins galectin-1, -3, -8 and -9 acts as a specific sensor of lysosomal damage by binding to the exposed glycosylated proteins Thurston et al. In addition, galectin-8 and galectin-9 can promote autophagy by inhibition of mTORC1 and activation of AMP-activated protein kinase AMPK , respectively Jia et al. In lysophagy, damaged lysosomes, as well as membrane remnants originating from complete lysosome rupture are engulfed by autophagosomes Papadopoulos and Meyer, Very recently, it has been reported that galectins-3, -8 and -9 coordinate a repair, removal and replacement program for damaged lysosomes. Here, galectin-3 recruits ESCRT components for repair of lysosomal membranes. When ESCRT-mediated lysosome repair fails, galectin-3 recruits TRIM16 for removal of unsalvageable lysosomes by autophagy, aided by the autophagy-inducing effects of galectin-8 and galectin-9 Jia et al. In this way, components of the endosomal system and the autophagy machinery are instrumental for the homeostasis of lysosomes. Autophagy and endocytosis mediate the degradation and recycling of intracellular and extracellular components, respectively. Both pathways involve the gradual maturation and fusions of vesicles, which traffic to their final destination, the lysosome. The emerging extensive crosstalk between these two pathways is therefore not surprising. Many questions regarding the identity and composition of various intracellular membrane compartments, as well as differential control of their intersections and the protein complexes involved still remain unanswered. Novel high-resolution live-cell imaging approaches combined with proteomics and CRISPR-based screens will undoubtedly provide further insight. Elucidation of the dynamic interplay between autophagy and endocytosis in the regulation of cell signaling is likely to provide exciting avenues for development of new therapeutic approaches. We thank members of the Molecular Cancer Research Group for important discussions and Trond Lamark for critical reading of the manuscript. Research in T. is supported by a grant from the Northern Norway Regional Health Authority Helse Nord RHF; grant number HNF We are now welcoming submissions for our upcoming Special Issue: Imaging Cell Architecture and Dynamics. This issue will be coordinated by two Guest Editors: Lucy Collinson The Francis Crick Institute, UK and Guillaume Jacquemet University of Turku, Finland. Submission deadline: 1 March People who know JCS well will know that we're more than just a journal and that our community — the cell biology community — really is at the heart of everything we do. Read the full Editorial by Editor-in-Chief Michael Way and Executive Editor Seema Grewal. Registration is open for our Journal Meeting Diversity and Evolution in Cell Biology, which aims to bring together evolutionary biologists and cell biologists investigating diverse aspects of cellular physiology. Submit your abstract by 5 April. Final registration deadline: 3 May Early-career researchers interested in the roles of nuclear lipids, apply now for one of the ten funded places at this Workshop, which will take place October Application deadline: 19 April. There are many benefits to publishing in Journal of Cell Science - read more about why you should choose JCS or visit our submission page now. Sign In or Create an Account. Search Dropdown Menu. header search search input Search input auto suggest. filter your search All content All journals Journal of Cell Science. Advanced Search. User Tools Dropdown. Sign in. Toggle Menu Menu Articles Accepted manuscripts Latest complete issue Issue archive Archive by article type Special issues Subject collections Interviews Essay series Sign up for alerts About us About JCS Editors and Board Editor biographies Travelling Fellowships Grants and funding Journal Meetings Workshops The Company of Biologists The Forest of Biologists Journal news For authors Submit a manuscript Aims and scope Presubmission enquiries Fast-track manuscripts Article types Manuscript preparation Cover suggestions Editorial process Promoting your paper Open Access JCS Prize Manuscript transfer network Biology Open transfer Journal info Journal policies Rights and permissions Media policies Reviewer guide Sign up for alerts For librarians Contacts Contacts Subscriptions Advertising Feedback. Skip Nav Destination Close navigation menu Article navigation. Volume , Issue Previous Article Next Article. Article contents. Overview of the autophagy pathway. Overview of endocytosis. Impact of autophagy on endocytosis and signaling. Autophagosome formation and its intersection with the endosomal system. Autophagosome maturation is dependent on fusion with the endolysosomal system. Vesicle transport governs fusion between autophagosomes and the endolysosomal system. Lysosome quality control. Conclusions and perspectives. Article Navigation. REVIEW 22 May Autophagy and endocytosis — interconnections and interdependencies In collection: Autophagy , Membrane Trafficking. Birgisdottir birgisdottir uit. no ; terje. johansen uit. This site. Google Scholar. Terje Johansen Author and article information. Competing interests The authors declare no competing or financial interests. Online ISSN: Norges Forskningsråd Kreftforeningen Helse Nord RHF HNF Published by The Company of Biologists Ltd. J Cell Sci 10 : jcs Cite Icon Cite. toolbar search Search Dropdown Menu. toolbar search search input Search input auto suggest. View large Download slide. Table 1. View Large. Box 1. LC3-associated phagocytosis and LAP-like processes. Box 2. Nutrient sensing and mTORC1 regulation to maintain lysosome homeostasis and autophagic flux. Funding Research in T. Search ADS. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Endolysosomes are the principal intracellular sites of acid hydrolase activity. Beyond self-eating: the control of nonautophagic functions and signaling pathways by autophagy-related proteins. TRIMs and galectins globally cooperate and TRIM16 and galectin-3 co-direct autophagy in endomembrane damage homeostasis. A mammalian autophagosome maturation mechanism mediated by TECPR1 and the AtgAtg5 conjugate. Pacer mediates the function of class III PI3K and HOPS complexes in autophagosome maturation by engaging Stx Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Identification of an adaptor-associated kinase, AAK1, as a regulator of clathrin-mediated endocytosis. To degrade or not to degrade: mechanisms and significance of endocytic recycling. De Duve. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. RAB2 regulates the formation of autophagosome and autolysosome in mammalian cells. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting AtgL1. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. Targeting of early endosomes by autophagy facilitates EGFR recycling and signalling. LC3 binding to the scaffolding protein JIP1 regulates processive dynein-driven transport of autophagosomes. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. A syntaxin SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. Mammalian Atg8 proteins regulate lysosome and autolysosome biogenesis through SNAREs. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. A Rab5 endosomal pathway mediates Parkin-dependent mitochondrial clearance. LC3-associated endocytosis facilitates β-amyloid clearance and mitigates neurodegeneration in murine Alzheimer's disease. A novel AAK1 splice variant functions at multiple steps of the endocytic pathway. LC3-associated phagocytosis - the highway to hell for phagocytosed microbes. Nutrient-dependent mTORC1 association with the ULK1-AtgFIP complex required for autophagy. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. Pharmacological modulators of autophagy activate a parallel noncanonical pathway driving unconventional LC3 lipidation. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Starvation-induced MTMR13 and RAB21 activity regulates VAMP8 to promote autophagosome-lysosome fusion. Galectin-3 coordinates a cellular system for lysosomal repair and removal. Vesicular stomatitis virus is believed to be taken up by the autophagosome from the cytosol and translocated to the endosomes where detection takes place by a pattern recognition receptor called toll-like receptor 7 , detecting single stranded RNA. Following activation of the toll-like receptor, intracellular signaling cascades are initiated, leading to induction of interferon and other antiviral cytokines. A subset of viruses and bacteria subvert the autophagic pathway to promote their own replication. When galectin-8 binds to a damaged vacuole , it recruits an autophagy adaptor such as NDP52 leading to the formation of an autophagosome and bacterial degradation. Autophagy degrades damaged organelles, cell membranes and proteins, and insufficient autophagy is thought to be one of the main reasons for the accumulation of damaged cells and aging. One of the mechanisms of programmed cell death PCD is associated with the appearance of autophagosomes and depends on autophagy proteins. This form of cell death most likely corresponds to a process that has been morphologically defined as autophagic PCD. One question that constantly arises, however, is whether autophagic activity in dying cells is the cause of death or is actually an attempt to prevent it. Morphological and histochemical studies have not so far proved a causative relationship between the autophagic process and cell death. In fact, there have recently been strong arguments that autophagic activity in dying cells might actually be a survival mechanism. Autophagy is essential for basal homeostasis ; it is also extremely important in maintaining muscle homeostasis during physical exercise. A study of mice shows that autophagy is important for the ever-changing demands of their nutritional and energy needs, particularly through the metabolic pathways of protein catabolism. In a study conducted by the University of Texas Southwestern Medical Center in Dallas , mutant mice with a knock-in mutation of BCL2 phosphorylation sites to produce progeny that showed normal levels of basal autophagy yet were deficient in stress-induced autophagy were tested to challenge this theory. Results showed that when compared to a control group, these mice illustrated a decrease in endurance and an altered glucose metabolism during acute exercise. Another study demonstrated that skeletal muscle fibers of collagen VI in knockout mice showed signs of degeneration due to an insufficiency of autophagy which led to an accumulation of damaged mitochondria and excessive cell death. Both studies demonstrate that autophagy induction may contribute to the beneficial metabolic effects of exercise and that it is essential in the maintaining of muscle homeostasis during exercise, particularly in collagen VI fibers. Work at the Institute for Cell Biology, University of Bonn, showed that a certain type of autophagy, i. chaperone-assisted selective autophagy CASA , is induced in contracting muscles and is required for maintaining the muscle sarcomere under mechanical tension. This is necessary for maintaining muscle activity. Because autophagy decreases with age and age is a major risk factor for osteoarthritis , the role of autophagy in the development of this disease is suggested. Proteins involved in autophagy are reduced with age in both human and mouse articular cartilage. Cancer often occurs when several different pathways that regulate cell differentiation are disturbed. Autophagy plays an important role in cancer — both in protecting against cancer as well as potentially contributing to the growth of cancer. The role of autophagy in cancer is one that has been highly researched and reviewed. There is evidence that emphasizes the role of autophagy as both a tumor suppressor and a factor in tumor cell survival. Recent research has shown, however, that autophagy is more likely to be used as a tumor suppressor according to several models. Several experiments have been done with mice and varying Beclin1, a protein that regulates autophagy. In support of the possibility that Beclin1 affects cancer development through an autophagy-independent pathway is the fact that core autophagy factors which are not known to affect other cellular processes and are definitely not known to affect cell proliferation and cell death, such as Atg7 or Atg5, show a much different phenotype when the respective gene is knocked out, which does not include tumor formation. In addition, full knockout of Beclin1 is embryonic lethal whereas knockout of Atg7 or Atg5 is not. Necrosis and chronic inflammation also has been shown to be limited through autophagy which helps protect against the formation of tumor cells. Cells that undergo an extreme amount of stress experience cell death either through apoptosis or necrosis. Prolonged autophagy activation leads to a high turnover rate of proteins and organelles. A high rate above the survival threshold may kill cancer cells with a high apoptotic threshold. Alternatively, autophagy has also been shown to play a large role in tumor cell survival. In cancerous cells, autophagy is used as a way to deal with stress on the cell. These metabolic stresses include hypoxia, nutrient deprivation, and an increase in proliferation. These stresses activate autophagy in order to recycle ATP and maintain survival of the cancerous cells. By inhibiting autophagy genes in these tumors cells, regression of the tumor and extended survival of the organs affected by the tumors were found. Furthermore, inhibition of autophagy has also been shown to enhance the effectiveness of anticancer therapies. New developments in research have found that targeted autophagy may be a viable therapeutic solution in fighting cancer. As discussed above, autophagy plays both a role in tumor suppression and tumor cell survival. Thus, the qualities of autophagy can be used as a strategy for cancer prevention. The first strategy is to induce autophagy and enhance its tumor suppression attributes. The second strategy is to inhibit autophagy and thus induce apoptosis. The first strategy has been tested by looking at dose-response anti-tumor effects during autophagy-induced therapies. These therapies have shown that autophagy increases in a dose-dependent manner. This is directly related to the growth of cancer cells in a dose-dependent manner as well. Secondly, inhibiting the protein pathways directly known to induce autophagy may also serve as an anticancer therapy. The second strategy is based on the idea that autophagy is a protein degradation system used to maintain homeostasis and the findings that inhibition of autophagy often leads to apoptosis. Inhibition of autophagy is riskier as it may lead to cell survival instead of the desired cell death. Negative regulators of autophagy, such as mTOR , cFLIP , EGFR , GAPR-1 , and Rubicon are orchestrated to function within different stages of the autophagy cascade. The end-products of autophagic digestion may also serve as a negative-feedback regulatory mechanism to stop prolonged activity. Regulators of autophagy control regulators of inflammation, and vice versa. Parkinson's disease is a neurodegenerative disorder partially caused by the cell death of brain and brain stem cells in many nuclei like the substantia nigra. Parkinson's disease is characterized by inclusions of a protein called alpha-synuclien Lewy bodies in affected neurons that cells cannot break down. Deregulation of the autophagy pathway and mutation of alleles regulating autophagy are believed to cause neurodegenerative diseases. Mutations of synuclein alleles lead to lysosome pH increase and hydrolase inhibition. As a result, lysosomes degradative capacity is decreased. There are several genetic mutations implicated in the disease, including loss of function PINK1 [] and Parkin. Mitochondria is involved in Parkinson's disease. In idiopathic Parkinson's disease, the disease is commonly caused by dysfunctional mitochondria, cellular oxidative stress, autophagic alterations and the aggregation of proteins. These can lead to mitochondrial swelling and depolarization. Excessive activity of the crinophagy form of autophagy in the insulin-producing beta cells of the pancreas could reduce the quantity of insulin available for secretion, leading to type 2 diabetes. Since dysregulation of autophagy is involved in the pathogenesis of a broad range of diseases, great efforts are invested to identify and characterize small synthetic or natural molecules that can regulate it. Contents move to sidebar hide. Article Talk. Read Edit View history. Tools Tools. What links here Related changes Upload file Special pages Permanent link Page information Cite this page Get shortened URL Download QR code Wikidata item. Download as PDF Printable version. In other projects. Wikimedia Commons. Cellular catabolic process in which cells digest parts of their own cytoplasm. Not to be confused with Autophagia. This article is about the cellular process. For other uses, see Autophagy disambiguation. Apoptosis — Programmed cell death in multicellular organisms Autophagy database Autophagin — Protease Pages displaying short descriptions with no spaces Mitophagy — autophagic process in which mitochondria are delivered to the vacuole and degraded Pages displaying wikidata descriptions as a fallback Residual body — vesicles containing indigestible materials, part of lysosomal digestion Pages displaying wikidata descriptions as a fallback Sub-lethal damage — Damaging changes to a biological cell Pages displaying short descriptions of redirect targets. A Greek—English Lexicon. Retrieved 6 September doi : PMID S2CID Journal of Molecular Biology. Nature Reviews. PMC Autophagy in health and disease. Progress in Molecular Biology and Translational Science. ISBN ISSN Radiotherapy and Oncology. Annual Review of Cell and Developmental Biology. Nature Cell Biology. Anselmier who first used the term "autophagie" in ". The Journal of Cell Biology. FEBS Letters. The Nobel Foundation. Retrieved 3 October The American Journal of Pathology. European Journal of Biochemistry. In Wang HG ed. Autophagy and Cancer. The Journal of Biological Chemistry. Proceedings of the National Academy of Sciences of the United States of America. Bibcode : PNAS Developmental Cell. Bibcode : Natur. Gordon Research Conference. Keystone Symposia on Molecular and Cellular Biology. Archived from the original on Retrieved The Biochemical Journal. Cell Structure and Function. Nature Reviews Molecular Cell Biology. Biological Chemistry. Cell Death and Differentiation. International Journal of Cell Biology. Nature Plants. Experimental Cell Research. Autophagy Database. Nature Education. Molecular and Cellular Biology. Biochimica et Biophysica Acta BBA - Molecular and Cell Biology of Lipids. Journal of Experimental Botany. Molecular Biology of the Cell. |
| Machinery, regulation and pathophysiological implications of autophagosome maturation | Autophagy is a general term for the degradation of cytoplasmic components within lysosomes Cuervo ; Levine and Klionsky ; Shintani and Klionsky ; Klionsky , ; Mizushima and Klionsky The IAP family member BRUCE regulates autophagosome-lysosome fusion. Article CAS PubMed PubMed Central Google Scholar Shen, K. Cell membrane Nucleus Endoplasmic reticulum Golgi apparatus Parenthesome Autophagosome Vesicle Exosome Lysosome Endosome Phagosome Vacuole Acrosome Cytoplasmic granule Melanosome Microbody Glyoxysome Peroxisome Weibel—Palade body. A family of proteins with 4—12 repeats of a β-stranded blade that function as structural scaffolds for ligand binding, enzymatic activity and assembly of multiple protein complexes. Inhibitors and activators of autophagosome formation, including VPS34 and ULK1 inhibitors or the activating peptide Tat—beclin 1 , are potent modulators of autophagy, but are not yet available for clinical use , |
der Interessante Moment
wacker, welche ausgezeichnete Mitteilung