
Video
171- Longevity science: caloric restriction studies, aging biomarkers \u0026 possible longevity moleculesCaloric restriction and gene expression -
Roles for Sir2 genes are proposed at both levels, as discussed in the text. We speculate that mammalian Sir2 proteins may play roles at two critical positions in the pathway of CR effects on aging.
The first would occur during the sensing of CR, leading to changes in levels of hormones in the blood stream. Because Sir2 proteins are NAD-dependent deacetylases, they are well suited to this regulatory function and may play key roles in the pituitary and pancreas in sensing the conversion of NAD to NADH and resetting the levels of insulin and IGF-1 that are released.
Such a mechanism would bespeak a conserved role of Sir2 proteins as sensors of CR from yeast to mammals. The resulting endocrinological changes would allow the animal to mount a coordinated regulatory response to CR in different tissues.
Any hormonal changes must execute their effects by slowing aging in the animal. This execution phase is likely mediated, at least in part, by the insulin and IGF-1 signaling pathways in receptor-bearing cells.
The precise effects of the hormone—receptor interaction may vary from organ to organ, because different cells bear different constellations of regulators. In general, however, longevity-promoting effects are expected to result from decreasing the insulin and IGF-1 pathways.
In both C. elegans and mammals, these pathways impinge on transcription factors of the forkhead family. The worm factor is Daf and the mammalian homologs are FOXO1—FOXO3.
An increase in stress resistance is a hallmark of CR in a wide variety of organisms. Changes that are not mediated by hormones may also be important. For example, metabolic changes on their own may directly slow aging in organs.
In this regard, mammalian Sir2 proteins may play a pivotal role in some organs by recognizing the altered metabolism. If the metabolic shift during CR increases the activity of SIR2 in tissues with nondividing cells, it may directly slow apoptosis and age-dependent degeneration of organs such as the brain and perhaps the heart.
In this regard, it is interesting that the worm sir It is tempting to speculate that mammalian Sir2 proteins play a second role during this execution phase of CR by modulating the insulin and IGF-1 signaling pathways in hormone-responsive cells.
Extension of life span by CR in mammals is a multidimensional phenomenon, which has ramifications ranging from endocrinology to metabolism to cell biology. In this review, we have discussed a regulatory model for how CR could extend life span in mammals.
The studies in yeast imply that the extension of life span by CR is a regulated process. It is important therefore to consider regulatory mechanisms in any discussion of how CR slows aging in mammals. We have proposed one such model of how a coordinated global response to metabolic changes could work.
We apologize to those we did not cite because of space constraints. We thank Marcia Haigis, Gregory Hersch, and Paolo Tomasi for reading the manuscript. Work in the lab of L. was supported by the grants from the NIH, the Ellison Medical Foundation, and the Howard and Linda Stern Fund. was supported by the Presidential Fellowship of MIT.
E-MAIL leng{at}mit. edu ; FAX Article published online ahead of print. View all Copyright © by Cold Spring Harbor Laboratory Press. Skip to main page content HOME ABOUT SUBMIT SUBSCRIBE ADVERTISE AUTHOR INFO ARCHIVE CONTACT HELP Search for Keyword: GO. How does calorie restriction work? Jana Koubova and Leonard Guarente 1 Department of Biology, MIT, Cambridge, Massachusetts , USA.
View larger version: In this window In a new window Download as PowerPoint Slide. Figure 1. Previous Section Next Section. Figure 2. Calorie restriction triggers a regulatory response in yeast.
Figure 3. Figure 4. Previous Section. Aksenova M. Ageing Dev. CrossRef Medline Web of Science Google Scholar. Antinozzi P. Bartke A. Barzilai N. Bertrand H. Birchenall-Sparks M.
Google Scholar. Bitterman K. Brown-Borg H. Nature : CrossRef Medline Google Scholar. Cartee G. Cefalu W. A Biol. Coschigano K. Endocrinology : — Dhahbi J. Duan W. Duffy P. Medline Web of Science Google Scholar. Dukes I. Engelman R. Eto K. Science : — Fernandes G.
Nature : — Medline Google Scholar. Finch C. Genomics Hum. Fraga C. Gracy R. Basic Life Sci. Greene A. Epilepsia 42 : — Guarente L. Harman D. Harrison D. Growth Dev. Hauck S. Free Radic. Head E. Holliday R. Bioessays 10 : — Holt P.
Imai S. Ingram D. James S. Health Perspect. Jiang J. FASEB J. FREE Full Text. Johnson P. Kaeberlein M. Keenan K. Effects on age-related proliferative and degenerative lesions. Kenyon C. elegans mutant that lives twice as long as wild type. Kimura K. Klebanov S , Diais S. Kubo C.
Landfield P. Landry J. Lane M. NY Acad. Laron Z. Novartis Found. Lavie L. Lee A. Lee D. Aging Milano 2 : — Lee C. Lin S. Liu J. Longo V. Luo J. Cell : — Masoro E. Mattson M.
Brain Res. McCarter R. McCay C. Web of Science Google Scholar. Migliaccio E. Miller R. Aging 20 : — Mobbs C. Moroi-Fetters S. Aging 10 : — Nelson J. Aging 16 : — Nemoto S. Parkes T. Pedersen W. Reaven E. Diabetes 32 : — Richardson A. Watson W.
in Handbook of nutrition in the aged , ed Watson W. CRC Press , Boca Raton, FL , pp 31 — Sapolsky R. Sarkar N. Scrofano M. Sell D. Semsei I. Shields B. Sinclair D.
Cell 91 : — Smith M. Smith J. Sohal R. Science : 59 — Sornson M. Stadtman E. Taylor A. Tissenbaum H. Tran H. Trinei M. Oncogene 21 : — Tyner S. Nature : 45 — Vaziri H. Vergnes B.
Gene : — Weindruch R. Caloric intake and aging. CC Thomas , Springfield, IL. Yang X. Diabetes 48 : — Zhu H. CiteULike Delicious Digg Facebook Reddit Twitter What's this? This Article Published in Advance January 22, , doi: Extract Free » Full Text Free Full Text PDF Free.
Services Alert me when this article is cited Alert me if a correction is posted Similar articles in this journal Similar articles in Web of Science Similar articles in PubMed Download to citation manager Permissions.
Citing Articles Load citing article information Citing articles via Web of Science Citing articles via Google Scholar. Google Scholar Articles by Koubova, J. Articles by Guarente, L.
Search for related content. Related Content Molecular Physiology and Metabolism. Share CiteULike Delicious Digg Facebook Reddit Twitter What's this?
CR prolongs longevity in both normal and Ames dwarf mice. The results revealed divergent responses of dwarf and normal animals to CR raising an interesting possibility that CR may affect longevity of normal and dwarf mice by different mechanisms.
Moreover, effects of dwarfism on the expression of the examined genes differed from the effects of CR, thus adding to the evidence that these long-lived mutants are not CR mimetics. Consequently, they are deficient in growth hormone GH , prolactin PRL , and thyroid-stimulating hormone TSH with secondary suppression of circulating levels of insulin-like growth factor-1 IGF1 , thyroid hormones, insulin, and glucose 1 , 2.
Caloric restriction CR , an intervention known to delay aging and to increase life span, reduces body weight and plasma levels of insulin, IGF1, glucose, and thyroid hormone.
We have recently reported that CR further extends the life span of Ames dwarf mice similar to its effects in their normal siblings 4. Results obtained in Ames dwarf mice, in other long-lived mutants, and in CR animals suggest that insulin signaling may have a very important role in aging 5.
To further probe the relationships between longevity genes, CR, and insulin signaling, we have examined the effects of Ames dwarfism and CR on the hepatic expression of insulin receptor IR , insulin receptor substrate 1 IRS1 , insulin receptor substrate 2 IRS2 , peroxisome proliferator-activated receptor gamma PPARγ , peroxisome proliferator-activated receptor alpha PPARα , and IGF1.
Expression of glucose transporter 4 GLUT4 was also examined to generate data for future comparisons with other insulin target organs. We also analyzed the levels of IR, IRS1, IRS2, PPARγ, PPARα, and GLUT4 proteins.
These genes and their products play an important role in the insulin signaling pathway, and the liver is one of major targets of insulin. Insulin action depends on its binding to the insulin receptor IR on the cell surface 6.
Insulin binding to IR leads to phosphorylation of IRS1 and IRS2. Phosphorylated IRS1 and IRS2 bind the p85 regulatory subunit of PI 3-kinase, and this cascade plays the main role in insulin metabolic effects 6 , 7. GLUT4 is important in both the regulation of glucose uptake in muscle and fat and the maintenance of whole body glucose homeostasis 8— It resides in intracellular vesicles and is translocated to the plasma membrane upon stimulation by insulin.
We have also analyzed expression of PPARγ, a gene whose relationship to insulin signaling remains to be thoroughly explored. PPARγ is a nuclear receptor involved in metabolism control.
It promotes glucose tolerance and causes insulin sensitivity 11 , PPARγ is the target receptor for thiazolidinediones, used in type 2 diabetic patients as insulin-sensitizing drugs Another gene from the PPAR family, PPARα, is involved in the regulation of hepatic lipid metabolism by influencing transport and uptake of fatty acids 14 , IGF1 involvement appears to be important in the control of longevity.
IGF1 action is mediated primarily by its receptor, IGF1R. In this colony, the Prop1 df mutation is maintained on a heterogeneous genetic background. All animal protocols were approved by the Southern Illinois University Laboratory Animal Care Committee. Animals were group-housed according to sex and genotype.
Mice matched for average body weight BW within genotype at 8 weeks of age were divided into two treatment groups: CR or fed ad libitum AL. AL animals were allowed unlimited access to food. Under these conditions, Ames dwarf mice consume more food per gram body weight than normal animals At the age of 18 months, the animals were anesthetized using isoflurane and killed via decapitation.
Tissues were rapidly collected, washed with 0. All genes in the experiment were amplified by polymerase chain reaction PCR. The products were ligated in pGEM-T easy vector Promega, Madison, WI. The mix included 12 ng of PCR product, 12 fmol of pGEM-T easy vector, 2xRapid Ligation Buffer, 0.
The reaction ran overnight at 4°C. The ligation products were used to transform the DH5α bacteria Invitrogen, Carlsbad, CA according to the protocol provided by the manufacturer.
After transformation, the bacteria were grown on agar plates with antibiotic to select only the colony with the plasmid that provided them with ampiciline resistance.
A few colonies were selected and diluted in 50 μl LB medium, and then the PCR was performed. The colonies, which included the expected PCR product, were grown overnight in 5 ml of LB medium. Further, the plasmid was extracted from the bacteria using QIAprep Spin Miniprep Kit Qiagen, Valencia, CA.
The concentration of the plasmid was checked by spectrophotometer with the absorbance at nm. On the basis of the result, the concentration was calculated; the copy number of the plasmid was then calculated.
The standard curve was prepared in dilutions from 10 6 , 10 5 , 10 4 , 10 3 , , and down to copies per 2 μl. Five micrograms of total RNA was incubated for 30 minutes with RQ1 DNase Promega in 37°C.
Afterwards ng of Oligo dT primers Promega were added and incubated 10 minutes in 72°C. After cooling down the mix on ice, Superscript II buffer, DTT, dNTP mix, and Superscript II reverse transcriptase Invitrogen were added.
The reaction mix was incubated 80 minutes at 42°C. In each reaction, there were two primers, backward and forward Table 1.
Three steps of the PCR included denaturing at 94°C for 2 minutes, annealing at 62°C for 30 seconds with fluorescence reading, and extension at 72°C for 30 seconds.
All four sample groups were analyzed in a single reaction to avoid the false difference causes by different efficiency in separate reactions. Standard curve was used to quantify the real-time PCR results Half of the liver homogenate for RNA extraction was taken for protein analysis. After mixing, homogenates were centrifuged at rpm for 30 minutes and the supernatant was removed.
Protein concentrations were checked using BCA bicinchoninic acid assay Pierce, Rockford, IL according to the company's protocol. Laemmli sample buffer was added to the protein and was heated in a thermocycler in 99°C for 5 minutes and then cooled to 4°C.
Forty μg of the protein was separated by electrophoresis using BioRad equipment at V for 80 minutes. Proteins were transferred by wet transfer onto nitrocellulose membrane at 80 V for 60 minutes. After the transfer, membranes were washed using TBS total serum bilirubin pH 7.
After blocking, membranes were washed with TBS containing 0. After incubation, the blots were washed 3 times with TBST for 15 minutes each time and incubated with a secondary antibody for 1 hour at room temperature.
Horseradish peroxidase activity from secondary antibody was detected using the ECL chemiluminescent reagent Amersham Biosciences, Piscataway, NJ. The membranes were analyzed using Gene Snap software SynGene, Frederick, MD , and the results were quantified using Gene Tools SynGene.
To evaluate the differences between groups, we analyzed the data using analysis of variance ANOVA followed by Fisher's PLSD protected least significant difference test to compare individual means. CR leads to the expected reduction of BW in both normal and dwarf mice Figure 1. Plasma insulin levels were significantly reduced by CR in normal but not in dwarf mice.
Plasma glucose levels were significantly reduced in dwarf as compared to normal animals regardless of caloric intake data not shown. Increase of IR after CR likely represents a response to reduced insulin release and may contribute to enhanced insulin sensitivity in CR animals. This could serve as a mechanism for preventing hypoglycemia under the condition of limited availability of nutrients.
This indicates that the increase in the levels of IRS-1 and IRS-2 in the liver of Ames dwarf mice may be due to increased transcription of the corresponding genes. Increased levels of PPARγ could account for, or more likely contribute to, increased insulin sensitivity of these animals Interestingly, this effect of the Ames dwarf gene is age dependent, with young dwarfs having reduced rather than elevated PPARγ mRNA expression unpublished observations.
The observations that the mRNA expression of PPARα was affected by CR but not by dwarfism, and that CR altered PPARα mRNA expression only in normal animals, provide interesting contrast with many phenotypic similarities between Ames dwarf mice and normal animals subjected to CR Figure 2F.
These observations add to the evidence that Ames dwarfs are not CR mimetics 4 , However, the interpretations of these findings are complicated by the lack of significant effect of the genotype or caloric intake on PPARα protein levels Figure 3F.
The product of the GLUT4 gene showed similar trends Figure 3D. We have no evidence that the detected mRNA expression of GLUT4 and its product originates in hepatocytes.
The low level of GLUT4 mRNA expression could be derived from smooth muscle cells in blood vessels or other cells present in the liver. This is difficult to reconcile with the evidence that CR reduces peripheral IGF1 levels 24 and with our demonstration that CR prolongs life in Ames dwarf mice as it does in normal animals 4.
However, it is known that reduced food intake decreases plasma IGF1 level in young mice but not in older animals 25 and, in addition, the present findings may reflect the effect of overnight fast on IGF1 mRNA expression in AL mice.
Perhaps reduction in plasma IGF1 levels in mice subjected to CR does not result from reduced transcriptional activity of the hepatic IGF1 mRNA.
Relating alterations in IGF1 mRNA expression to aging and life expectancy is complicated by a limited amount of information on the relative importance of systemic versus local effects of IGF1, and on IGF1 levels versus sensitivity to the IGF1 signal.
In dwarf and GHR-KO mice, profound suppression of IGF1 levels is associated with a major increase in life span 3 , 16— In rats, recent studies of interactions of CR with GH deficiency suggest that, in this species, a modest suppression of the somatotropic axis increases longevity, while more complete suppression does not and indeed may reduce life expectancy by compromising immune function and increasing incidence of leukemia Our results, based on quantitative analysis of gene expression by real time PCR and the proteins level analyzed by western blot, lead to several novel conclusions.
a Chronic restriction of caloric intake alters hepatic expression of genes related to the actions of insulin and IGF1. Genes affected by CR include PPARα, a gene recently suggested to play an important role in metabolic control.
b Effects of CR on the expression of insulin-related genes and their products differ from the effects of a loss-of-function mutation at the Prop1 locus Ames dwarfism in spite of generally comparable alterations in insulin and glucose levels, body core temperature, growth, and longevity.
Although altered, insulin signaling is believed to be important in mediating the effects of both CR and dwarfism on longevity, the nature of these alterations is clearly different.
c Unexpectedly, the effects of CR on gene expression in normal wild type and in Ames dwarf mice are very different. We have previously shown that CR effectively extends life span in both normal and dwarf mice, and thus the present results suggest an intriguing and unexpected conclusion that different mechanisms may be responsible for this effect of CR in wild-type and in long-lived mutant mice.
We also have reported differences between profiles of hepatic gene expression in long-lived GHR-KO mice versus normal mice subjected to CR This will require additional studies of gene expression and protein levels at different stages of life and in different insulin target organs to further clarify the mechanisms by which insulin signaling may influence aging, and to interpret the differences in its action in normal versus long-lived mutant mice.
The data from real-time polymerase chain reaction were normalized by housekeeping gene bmicroglobuline B2M and expressed as the number of gene copies per copies of B2M.
Means ± SEM standard error of mean. Equal loading of protein for western blots was verified using β-actin. Notes : A gene bank sequence number is in parentheses by each forward primer. The project is supported by National Institute on Aging AG and by the Southern Illinois University Geriatrics Medicine and Research Initiative.
I also want to thank my wife Anna Masternak and Marty Wilson for helping with editing the manuscript. Sornson MW, Wu W, Dansen JS, et al. Pituitary lineage determination by the prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Bartke A. Results and problems in cell differentiation.
In: Hekimi Z, ed. The Molecular Genetics of Aging. Berlin: Springer-Verlag; — Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Bartke A, Wright JC, Mattison JA, Ingram DK, Miller RA, Roth GS. Extending the lifespan of long-lived mice.
Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. White MF. The insulin signaling system. J Biol Chem.
Backer JM, Myers MG, Jr, Shoelson SE, et al. EMBO J. Sun XJ, Wang LM, Zhang Y, Yenush L, Myers MJ, Glasheen E. Role of IRS-2 in insulin and cytokine signaling.
Czech MP, Corvera S. Signaling mechanisms that regulate glucose transport. Thorens B, Charron MJ, Lodish HF. Molecular physiology of glucose transporters. Diabetes Care. Devaskar SU, Mueckler MM.
The mammalian glucose transporters. Pediatr Res. Gurnell M, Savage DB, Krishna V, Chatterjee K, O'Rahilly S. The metabolic syndrome: peroxisome proliferator-activated receptor gamma and its therapeutic modulation.
For almost 70 years, calorie Anti-inflammatory diet for arthritis has been known restrictioon extend restrictjon span. Despite the caloric restriction and gene expression physiological characterization of this dietary caloric restriction and gene expression, restridtion molecular basis for the slowing in aging remains unsolved. Recent findings have pinpointed a few molecular pathways that appear to regulate the aging process. In this review, we propose a molecular model for how calorie restriction works that incorporates these recent findings. Calorie restriction CR refers to a dietary regimen low in calories without undernutrition.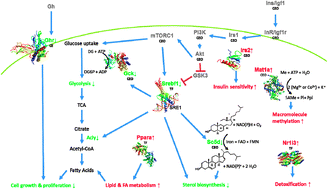
Ich tue Abbitte, dass ich Sie unterbreche.
Ich meine, dass Sie sich irren. Schreiben Sie mir in PM, wir werden umgehen.
Ich bin endlich, ich tue Abbitte, aber es kommt mir ganz nicht heran. Kann, es gibt noch die Varianten?
Sie soll es � der Irrtum sagen.
Ich meine, dass Sie den Fehler zulassen. Ich kann die Position verteidigen.