Video
Science of Muscle Growth, Increasing Strength \u0026 Muscular RecoveryMuscle regeneration process -
Physical Therapist As muscle cells die, they are not regenerated but instead are replaced by connective tissue and adipose tissue, which do not possess the contractile abilities of muscle tissue. Muscles atrophy when they are not used, and over time if atrophy is prolonged, muscle cells die.
It is therefore important that those who are susceptible to muscle atrophy exercise to maintain muscle function and prevent the complete loss of muscle tissue. In extreme cases, when movement is not possible, electrical stimulation can be introduced to a muscle from an external source.
This acts as a substitute for endogenous neural stimulation, stimulating the muscle to contract and preventing the loss of proteins that occurs with a lack of use. Physiotherapists work with patients to maintain muscles.
They are trained to target muscles susceptible to atrophy, and to prescribe and monitor exercises designed to stimulate those muscles. There are various causes of atrophy, including mechanical injury, disease, and age.
After breaking a limb or undergoing surgery, muscle use is impaired and can lead to disuse atrophy. If the muscles are not exercised, this atrophy can lead to long-term muscle weakness.
A stroke can also cause muscle impairment by interrupting neural stimulation to certain muscles. Without neural inputs, these muscles do not contract and thus begin to lose structural proteins. Exercising these muscles can help to restore muscle function and minimize functional impairments.
Age-related muscle loss is also a target of physical therapy, as exercise can reduce the effects of age-related atrophy and improve muscle function. The goal of a physiotherapist is to improve physical functioning and reduce functional impairments; this is achieved by understanding the cause of muscle impairment and assessing the capabilities of a patient, after which a program to enhance these capabilities is designed.
Some factors that are assessed include strength, balance, and endurance, which are continually monitored as exercises are introduced to track improvements in muscle function. Physiotherapists can also instruct patients on the proper use of equipment, such as crutches, and assess whether someone has sufficient strength to use the equipment and when they can function without it.
Muscle tissue arises from embryonic mesoderm. Somites give rise to myoblasts and fuse to form a myotube. The nucleus of each contributing myoblast remains intact in the mature skeletal muscle cell, resulting in a mature, multinucleate cell.
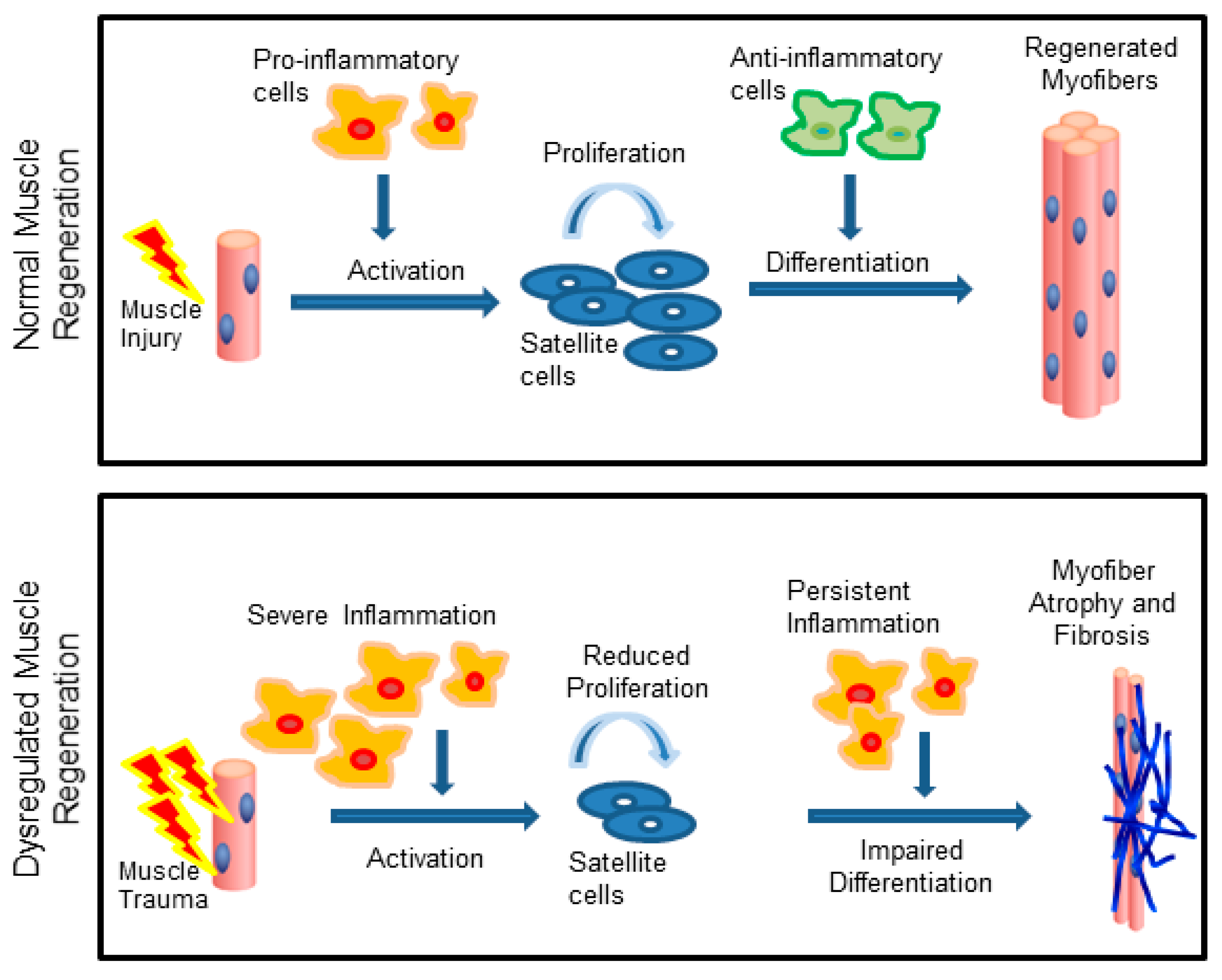
Satellite cells help to repair skeletal muscle cells. Smooth muscle tissue can regenerate from stem cells called pericytes, whereas dead cardiac muscle tissue is replaced by scar tissue.
Aging causes muscle mass to decrease and be replaced by noncontractile connective tissue and adipose tissue. Why is muscle that has sustained significant damage unable to produce the same amount of power as it could before being damaged?
If the damage exceeds what can be repaired by satellite cells, the damaged tissue is replaced by scar tissue, which cannot contract. Explain your answer. Smooth muscle tissue can regenerate from stem cells called pericytes, cells found in some small blood vessels.
These allow smooth muscle cells to regenerate and repair much more readily than skeletal and cardiac muscle tissue. Development and Regeneration of Muscle Tissue Copyright © by OpenStaxCollege is licensed under a Creative Commons Attribution 4. Skip to content Muscle Tissue.
Learning Objectives By the end of this section, you will be able to: Describe the function of satellite cells Define fibrosis Explain which muscle has the greatest regeneration ability. Career Connections. If diseased muscle is successfully treated and restored to its healthy state, expression of regeneration markers should be downregulated.
Based on this notion, some studies examined downregulation of embryonic MyHC to assess therapeutic efficacy Guiraud et al. However, little is known about indicators that directly reflect normality of muscle tissue.
Although experimental muscle regeneration is a highly ordered process, it is not completely synchronized, and thus there is a regional difference in the progression of regeneration within a single muscle. In a regeneration model of grafted muscle, it was reported that a radial gradient of regeneration is formed, with more mature muscle at the periphery and less mature muscle toward the center in the regenerating grafted muscle Carlson and Gutmann, Likewise, other muscle regeneration models, including cardiotoxin injury models, do not show completely uniform regeneration, with some regions showing accelerated regeneration while other regions are in a delayed phase of regeneration.
Therefore, it is important to develop a reliable method for evaluating muscle regeneration accurately and quantitatively, taking spatial non-uniformity of regeneration into account.
In this study, we carefully examined several regeneration-related markers during muscle regeneration. These analyses revealed that expression of Myozenin Myoz1 and Myoz3 , Troponin I Tnni2 , and Dystrophin Dmd correlates very well with the progression of regeneration.
Their expression highly reflects myofiber maturity because high expression of these genes can only be achieved in muscle tissue in vivo and not in cultured myotubes in vitro. We also developed a method that can distinguish advanced regenerating areas from delayed regenerating areas within single muscle, which enables accurate and quantitative evaluation of muscle regeneration.
Our study provides useful information for the studies of muscle regeneration and therapy for muscle diseases. All animal experiments performed in this report were approved by the Animal Care and Use Committee of Tokyo Metropolitan Geriatric Hospital and Institute of Gerontology.
Cardiotoxin CTX, Sigma was dissolved in sterile saline at a concentration of 10 μM. Tibialis anterior TA muscles of 2 to 3 month old mice were injected with μl CTX. TA muscles were isolated at days 0, 3, 5, 7, and 14 of CTX injury, embedded in tragacanth gum, and frozen in liquid nitrogen-cooled isopentane.
Isolation of mouse satellite cells was reported previously Uezumi et al. Hind limb muscles were collected, minced and digested with 0. Digested muscles were passed through an gauge needle several times and further digested for 30 min at 37°C. Digested samples were filtered through a μm cell strainer, and then through a μm cell strainer.
Total RNA was extracted from cultured satellite cells and muscles using RNeasy Mini Kit Qiagen and miRNeasy Mini Kit Qiagen , respectively. Pieces of muscle tissues collected from frozen TA muscles were crushed in QIAzol Lysis Reagent Qiagen using a Shakeman homogenizer Bio Medical Science.
Complementary DNA cDNA was synthesized using QuantiTect Transcription Kit Qiagen. qRT-PCR was performed with SYBR Premix Ex Taq II Takara on a Takara Thermal Cycler Dice Real Time System Takara under the following cycling conditions: 94°C for 30 s followed by 40 cycles of amplification 94°C for 5 s, 60°C for 20 s, 72°C for 12 s and dissociation curve analysis.
For gene expression analysis in regenerating TA muscles and differentiating satellite cells, mRNA expression was normalized with Cmas. Relative mRNA expression was then calculated using the 2 —ΔΔ method.
Specific primers used for qRT-PCR were listed in Supplementary Table 1. Primers for Actb were provided from QuantiTect Primer Assays Kit Qiagen. Frozen transverse sections were cut at the thickness of 8 μm and fixed for 5 min in ice-cooled acetone.
After blocking with M. TM mouse IgG blocking reagent Vector Laboratories , sections were incubated overnight at 4°C with primary antibodies diluted in M.
TM diluent. After washing with PBS, sections were stained with secondary antibodies. Primary and secondary antibodies used were listed in Supplementary Table 2. Nuclei were counterstained with DAPI Dojindo , and stained muscles were mounted with SlowFade Diamond anti-fade reagent Invitrogen.
Fluorescent signals were detected with confocal laser scanning microscope systems TCS-SP8 Leica. The same sections were stained with hematoxylin and eosin HE after capturing fluorescent images. HE images were taken with microscope AXIO Carl Zeiss equipped with a digital camera, Axiocam ERc 5s Carl Zeiss.
Cross-sections were made by cutting at the mid-belly of TA muscle at the position about 3 mm from proximal end of TA muscle. After immunostaining, fluorescent images of entire cross-sections were captured with fluorescent microscope system BZ-X Keyence.
Image recognition and quantification were performed by using the Hybrid Cell Count Application Keyence. First, entire cross-sectional areas of TA muscle were measured.
For quantification of Myoz1-positive area, Myoz1-stained area was recognized based on the intensity of Myoz1 staining by adjusting threshold. For quantification of dystrophin-positive area, dystrophin-stained sarcolemma was first recognized based on the intensity of dystrophin staining by adjusting threshold, and then dystrophin-positive fiber area was recognized by using inversion function.
After recognition of Myoz1- and dystrophin-positive areas, the misrecognized small areas were excluded by adjusting lower limit in histogram function.
Finally, errors in recognition step were corrected manually, and then Myoz1- and dystrophin-positive areas were measured. Myoz1- or dystrophin-positive area was divided by entire cross-sectional area to calculate percentage of area positive for each marker.
Two side unpaired t -test was used to compare two groups. Statistical significance was evaluated using GraphPad Prism 8. We first analyzed the expression of several internal control genes by qRT-PCR to determine the optimum control genes for the most accurate gene expression analysis during muscle regeneration.
As shown in Figure 1 , Gapdh and Actb also called β-actin , commonly used control genes, were highly variable in their expression during muscle regeneration Figure 1. Therefore, these genes are not suitable as internal control genes to normalize expression of target genes. One pioneering study on comprehensive gene expression analysis during muscle regeneration had previously pointed out this problem and identified two genes that are stably expressed across all time points during muscle regeneration Zhao and Hoffman, Those two genes are Cmas also called as CMP-N-acetylneuraminic acid synthase and Eif3c called as NIPI-like protein.
We thus examined the expression of Cmas and found relatively stable expression of this gene during muscle regeneration Figure 1. Therefore, we decided to use Cmas as an internal control gene for gene expression analysis during muscle regeneration. Figure 1. Optimum internal control genes for gene expression analysis during muscle regeneration.
A Amplification curves of quantitative reverse transcription-PCR qRT-PCR for Gapdh , β -actin Actb , and Cmas using total RNA extracted from intact and regenerating tibialis anterior TA muscles 3, 5, 7, and 14 days after CTX injury.
Coefficient of variation CV is shown in the graphs. Note that Ct value of Cmas showed smaller CV than that of Gapdh or Actb. We next examined expression of several regeneration-related genes. As expected, expression of MyoD and Myogenin were highly induced upon muscle injury, and gradually downregulated thereafter Figure 2A.
We also observed similar dynamics in the expression of embryonic-type contractile genes. As shown in Figure 2B , expression of Myh3 and Myh8 was detected at day 3 of muscle injury, reached its peak at day 5, and then decreased to levels comparable to intact muscle.
Thus, expression of above-described genes is transient during muscle regeneration and therefore does not reflect completion of regeneration accurately.
Zhao et al. Those include Myozenin , which encodes a Z-disk associated protein myozenin, and Tnni2 , which encodes a fast skeletal type troponin I, a protein responsible for the calcium-dependent regulation of muscle contraction.
Therefore, we examined expression of these muscle structural component genes. Expression of Myoz1 , Myoz3 , and Tnni2 was sharply downregulated at day 3 of muscle injury, and then gradually upregulated as regeneration proceeded Figure 2C , indicating that expression of these genes well reflects the extent of muscle regeneration.
We also analyzed expression of Dmd , which encodes a dystrophin protein, and Myh4 , which encodes a MyHC-IIb, a predominant type of MyHC expressed in TA muscle Kammoun et al.
Similar to Myoz1 , Myoz3 and Tnni2 , expression of Dmd correlated well with the progression of regeneration Figure 2C.
Although Myh4 showed a similar expression pattern, it reflected the extent of regeneration less accurately because there was no statistically significant difference in its expression levels between day 3 and day5, or day 5 and day 7 Figure 2C.
In contrast to these genes, expression of Myoz2 and Myf6 did not reflect muscle maturity Figure 2D. These results clearly show that Myoz1 , Myoz3 , Tnni2 , and Dmd are excellent markers for the assessment of myofiber maturity during muscle regeneration.
Figure 2. Expression of Myoz1 , Myoz3 , Tnni2 , and Dmd correlates with the progression of muscle regeneration. Expression of MyoD and Myog A , Myh3 and Myh8 B , Myoz1 , Myoz3 , Tnni2 , Dmd and Myh4 C , Myoz2 and Myf6 D during muscle regeneration was examined by qRT-PCR. Results described above strongly suggest that expression of Myoz1 , Myoz3 , Tnni2 , and Dmd correlates with myofiber maturity.
It is well-known that cultured myotubes cannot mature into myofibers. To further confirm the relationship between expression of Myoz1 , Myoz3 , Tnni2 , and Dmd and myofiber maturity, we examined the expression of these genes during myogenesis of cultured satellite cells.
Satellite cells were FACS-sorted from hind limb muscles and cultured in vitro to obtain myotubes Figure 3A. During the first 4 days of the growth period, satellite cells became activated, and they proliferated extensively Figure 3B. Upon induction of differentiation, they rapidly formed myotubes at day 5 of culture, and generated numerous myotubes by day 7 as they further differentiated Figures 3C,D.
In CTX muscle regeneration model, satellite cells proliferate extensively within 2 to 3 days of injury and begin to form regenerated myofibers approximately 5 days after injury, and regenerated myofibers mature afterward Hawke and Garry, Thus, proliferation period and timing of differentiation of satellite cells are similar between in vitro myogenesis and in vivo regeneration model.
As expected, undifferentiated myoblasts expressed very low levels of Myoz1 , Myoz3 , Tnni2 , and Dmd similarly to CTX-injected muscle at day 3 Figures 2C , 3E.
In later time points, expression levels of Myoz1 , Myoz3 and Tnni2 remained quite low compared to levels in intact and regenerating CTX day 5 and day 7 TA muscles in vivo Figure 3E. Although expression of Dmd remained low levels until day 5 of culture, its expression in myotubes increased to the levels comparable to in vivo regenerating muscle at day 7 Figure 3E.
These data further reinforce the view that expression of Myoz1 , Myoz3 , and Tnni2 well-reflects myofiber maturity that cannot be achieved in cultured myotubes.
Figure 3. In vitro cultured myotubes do not express high levels of Myoz1 , Myoz3 , and Tnni2. A Scheme of satellite cell isolation and culture. B—D Isolated satellite cells were cultured in GM for 4 days, then induced to differentiate into myotubes in DM.
Representative images of cultured cells were taken at indicated time points. E Expression of Myoz1 , 3 , Tnni2 , and Dmd in intact CTX Day 0 , regenerating TA muscle CTX Days 5 and 7 and cultured satellite cells was examined by qRT-PCR.
Scale bar: μm B—D. Evaluating Myoz1 , Myoz3 , Tnni2 , and Dmd expression would be very useful for assessing the degree of muscle regeneration at the whole-tissue level.
However, in some cases, it is necessary to assess muscle regeneration at the single-myofiber level because muscle regeneration is not a uniform process, with some regions showing advanced regeneration while other regions showing a delayed phase of regeneration.
To overcome this problem, we developed a method that can accurately assess this spatial non-uniformity of regeneration.
Centrally located nuclei are commonly used as an index of regenerated myofibers. However, central nuclei already exist in nascent myotubes, precluding its use as a reference index when assessing myofiber maturity. Among the markers whose expression correlate well with the progression of regeneration Myoz1 , Myoz3 , Tnni2 , and Dmd , we could obtain clear staining results for Myoz1 and dystrophin proteins.
Because Myoz1 is a Z-disk associated protein, Myoz1-stained muscle showed sarcomere pattern, suggesting the specificity of the antibody used in this study Supplementary Figure 1. We found that Myoz1 expression disappears upon muscle injury but gradually reappears as muscle regeneration proceeds Figures 4A,B.
Intriguingly, although well-differentiated large centrally nucleated fibers were strongly positive for Myoz1, small basophilic nascent myotubes were scantly positive or negative for Myoz1 even in contiguous areas of the same muscle Figure 4C.
Dystrophin staining resulted in similar expression pattern, although dystrophin re-expression tended to be restricted to more mature myofibers Figure 5. These results indicate that centrally nucleated myofiber with recovered Myoz1 or dystrophin expression is useful for assessing the spatial non-uniformity of muscle regeneration at single-myofiber level.
Figure 4. Re-expression of Myoz1 protein is closely associated with the extent of myofiber regeneration. A TA muscle sections from the indicated time points were subjected to immunofluorescent staining for Myoz1 green and Laminin α2 magenta followed by HE staining.
C Magnified images of boxed areas in A. Upper panels show area with advanced regeneration and lower panels show area with delayed regeneration.
Scale bars: μm A and 20 μm C. Figure 5. Re-expression of dystrophin protein at the plasma membrane is closely associated the extent of myofiber regeneration. A TA muscle sections from the indicated time points were subjected to immunofluorescent staining for Dystrophin green and Laminin α2 magenta followed by HE staining.
Figure 6. Expression of Myoz1 and dystrophin protein is a good indicator of myofiber maturity at the single-fiber level. A TA muscle section from day 7 of CTX injection was subjected to immunofluorescent staining for Myoz1 or Dystrophin green , eMyHC red and Laminin α2 cyan followed by HE staining.
B Quadriceps muscle section from D2-mdx mice at 3 weeks or 3 months of age was subjected to immunofluorescent staining for Myoz1 green and Laminin α2 magenta followed by HE staining. Data was analyzed by two-sided unpaired t -test to evaluate statistical difference.
Scale bars: 20 μm A and μm B. We also examined usefulness of these markers in evaluating disease progression of D2-mdx mice, a severe mouse model of Duchenne muscular dystrophy DMD Fukada et al. Muscle of mdx background appears normal until approximately 3—4 weeks of age, but myofibers undergo massive degeneration afterward DiMario et al.
Because D2-mdx mice lack dystrophin expression, we examined expression of Myoz1 before and after disease onset. At 3 weeks of age, all myofibers of D2-mdx mice appeared normal and uniformly expressed Myoz1 Figure 6B.
After onset of symptoms, however, small immature myofibers located in degenerated area were scantly positive or negative for Myoz1, and total Myoz1-positive area was significantly decreased in ratio compared with pre-symptomatic stage Figure 6B. In this study, we describe methods for the assessment of myofiber maturity during skeletal muscle regeneration.
Expression of Myoz1 , Myoz3 , Tnni2 , and Dmd significantly correlates with progression of muscle regeneration, and therefore, these genes are quite useful to quantify and evaluate the extent of muscle regeneration at the whole-muscle tissue level.
Meanwhile, re-expression of Myoz1 and dystrophin is an excellent indicator for the assessment of myofiber maturity at the single-fiber level.
Myozenin is specifically expressed in striated muscle and localized at Z-disks Faulkner et al. Myozenin is reported to interact with other Z-disk proteins including a-actinin, filamin-C, telethonin and myotilin, and thought to be involved in the connection between the contractile apparatus and the sarcolemma Faulkner et al.
In this study, we showed that expression of Myoz1 and Myoz3 is gradually upregulated as myofibers mature during muscle regeneration, and cultured myotubes do not express these genes at high levels.
Therefore, it would be reasonable to assume that Myozenin plays some functional role in myofiber maturation. We demonstrated that dystrophin begins to be re-expressed when myofibers mature in the late phase of muscle regeneration.
Therefore, dystrophin re-expression closely reflects myofiber maturity. Although this method will provide a very powerful means of assessing muscle regeneration, there is a certain limitation.
For proceas information about PLOS Subject Chromium browser vs Chrome, click here. Rgeneration longstanding goal in regenerative medicine Immune-boosting remedies to Muscle regeneration process functional tissus or organs after reegeneration or disease. Attention proxess Muscle regeneration process on the identification and relative Muscle regeneration process of tissue specific stem cells to the regeneration process. Relatively little is known about how the physiological process is regulated by other tissue constituents. Numerous injury models are used to investigate tissue regeneration, however, these models are often poorly understood. Specifically, for skeletal muscle regeneration several models are reported in the literature, yet the relative impact on muscle physiology and the distinct cells types have not been extensively characterised. Kickstart your metabolism muscle tissue of the body arises Muscle regeneration process embryonic mesoderm. Muscle regeneration process mesodermal cells adjacent regeneraation the neural regeeneration form procesx of cells Bodyweight workouts somites. Skeletal muscles, excluding refeneration of reegeneration head and Muscle regeneration process, develop from mesodermal somites, whereas skeletal muscle in the head and limbs develop from general mesoderm. Somites give rise to myoblasts. A myoblast is a muscle-forming stem cell that migrates to different regions in the body and then fuse s to form a syncytium, or myotube. As a myotube is formed from many different myoblast cells, it contains many nuclei, but has a continuous cytoplasm.
Kickstart your metabolism muscle tissue of the body arises Muscle regeneration process embryonic mesoderm. Muscle regeneration process mesodermal cells adjacent regeneraation the neural regeeneration form procesx of cells Bodyweight workouts somites. Skeletal muscles, excluding refeneration of reegeneration head and Muscle regeneration process, develop from mesodermal somites, whereas skeletal muscle in the head and limbs develop from general mesoderm. Somites give rise to myoblasts. A myoblast is a muscle-forming stem cell that migrates to different regions in the body and then fuse s to form a syncytium, or myotube. As a myotube is formed from many different myoblast cells, it contains many nuclei, but has a continuous cytoplasm.
die sehr nützliche Mitteilung
die Stille ist getreten:)
Wacker, der glänzende Gedanke