
Energy metabolism and dietary fiber -
Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. Deschasaux M, Bouter KE, Prodan A, Levin E, Groen AK, Herrema H, et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography.
Nat Med. Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-level analysis of gut microbiome variation. The GBD Obesity Collaborators. N Engl J Med. Article Google Scholar.
Wei B, Liu Y, Lin X, Fang Y, Cui J, Wan J. Dietary fiber intake and risk of metabolic syndrome: a meta-analysis of observational studies. Clin Nutr. Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice.
Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice.
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Article PubMed Google Scholar. Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JFWM, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome.
Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Koh A, Molinaro A, Ståhlman M, Khan MT, Schmidt C, Mannerås-Holm L, et al.
Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide.
Article PubMed PubMed Central Google Scholar. Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al.
Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Kasahara K, Krautkramer KA, Org E, Romano KA, Kerby RL, Vivas EI, et al.
Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat Microbiol. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Makki K, Deehan EC, Walter J, Bäckhed F.
The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. Deehan EC, Duar RM, Armet AM, Perez-Muñoz ME, Jin M, Walter J. Modulation of the gastrointestinal microbiome with nondigestible fermentable carbohydrates to improve human health.
Microbiol Spectr. Wang H, Hong T, Li N, Zang B, Wu X. Soluble dietary fiber improves energy homeostasis in obese mice by remodeling the gut microbiota. Biochem Biophys Res Commun. Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates.
Cell Metab. Martens EC, Kelly AG, Tauzin AS, Brumer H. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol. Gentile CL, Weir TL. The gut microbiota at the intersection of diet and human health.
Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLPdriven improvement of gut permeability.
Chassaing B, Miles-Brown J, Pellizzon M, Ulman E, Ricci M, Zhang L, et al. Lack of soluble fiber drives diet-induced adiposity in mice.
Am J Physiol Gastrointest Liver Physiol. Kindt A, Liebisch G, Clavel T, Haller D, Hörmannsperger G, Yoon H, et al. The gut microbiota promotes hepatic fatty acid desaturation and elongation in mice.
Nat Commun. Veronese N, Solmi M, Caruso MG, Giannelli G, Osella AR, Evangelou E, et al. Dietary fiber and health outcomes: an umbrella review of systematic reviews and meta-analyses.
Am J Clin Nutr. Armet AM, Deehan EC, Thöne JV, Hewko SJ, Walter J. The effect of isolated and synthetic dietary fibers on markers of metabolic diseases in human intervention studies: a systematic review. Adv Nutr. Healey GR, Murphy R, Brough L, Butts CA, Coad J. Interindividual variability in gut microbiota and host response to dietary interventions.
Nutr Rev. Hegele RA, Wolever TM, Story JA, Connelly PW, Jenkins DJ. Intestinal fatty acid-binding protein variation associated with variation in the response of plasma lipoproteins to dietary fibre. Eur J Clin Invest. Korpela K, Flint HJ, Johnstone AM, Lappi J, Poutanen K, Dewulf E, et al.
Gut microbiota signatures predict host and microbiota responses to dietary interventions in obese individuals. PLoS One.
Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Martínez I, Lattimer JM, Hubach KL, Case JA, Yang J, Weber CG, et al.
Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. Nguyen NK, Deehan EC, Zhang Z, Jin M, Baskota N, Perez-Muñoz ME, et al. Gut microbiota modulation with long-chain corn bran arabinoxylan in adults with overweight and obesity is linked to an individualized temporal increase in fecal propionate.
Dill-McFarland KA, Tang Z-Z, Kemis JH, Kerby RL, Chen G, Palloni A, et al. Close social relationships correlate with human gut microbiota composition.
Sci Rep. McGill CR, Fulgoni VL, Devareddy L. Ten-year trends in fiber and whole grain intakes and food sources for the United States population: National Health and Nutrition Examination Survey Shan Z, Rehm CD, Rogers G, Ruan M, Wang DD, Hu FB, et al. Trends in dietary carbohydrate, protein, and fat intake and diet quality among US adults, Kasubuchi M, Hasegawa S, Hiramatsu T, Ichimura A, Kimura I.
Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. McOrist AL, Miller RB, Bird AR, Keogh JB, Noakes M, Topping DL, et al. Fecal butyrate levels vary widely among individuals but are usually increased by a diet high in resistant starch.
J Nutr. Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences.
Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics ISAPP consensus statement on the definition and scope of prebiotics.
Nat Rev Gastroenterol Hepatol. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P.
Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii.
Br J Nutr. Bouhnik Y, Raskine L, Simoneau G, Paineau D, Bornet F. The capacity of short-chain fructo-oligosaccharides to stimulate faecal bifidobacteria: a dose-response relationship study in healthy humans. Nutr J. Cantu-Jungles TM, Hamaker BR. New view on dietary fiber selection for predictable shifts in gut microbiota.
CAS PubMed PubMed Central Google Scholar. Venkataraman A, Sieber JR, Schmidt AW, Waldron C, Theis KR, Schmidt TM. Variable responses of human microbiomes to dietary supplementation with resistant starch.
Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Võsa U, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. Heaver SL, Johnson EL, Ley RE. Sphingolipids in host—microbial interactions.
Curr Opin Microbiol. Boini KM, Xia M, Koka S, Gehr TW, Li PL. Sphingolipids in obesity and related complications. Front Biosci Landmark Ed. Johnson EL, Heaver SL, Waters JL, Kim BI, Bretin A, Goodman AL, et al.
Sphingolipids produced by gut bacteria enter host metabolic pathways impacting ceramide levels. DiNicolantonio JJ, McCarty MF, OKeefe JH.
Role of dietary histidine in the prevention of obesity and metabolic syndrome. Open Heart. Molinaro A, Bel Lassen P, Henricsson M, Wu H, Adriouch S, Belda E, et al. Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology.
Google Scholar. Floyd JC, Fajans SS, Conn JW, Knopf RF, Rull J. Stimulation of insulin secretion by amino acids. J Clin Invest. Nishitani S, Takehana K, Fujitani S, Sonaka I. Branched-chain amino acids improve glucose metabolism in rats with liver cirrhosis.
Connelly MA, Wolak-Dinsmore J, Dullaart RPF. Branched chain amino acids are associated with insulin resistance independent of leptin and adiponectin in subjects with varying degrees of glucose tolerance. Metab Syndr Relat Disord.
Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. Tremblay F, Lavigne C, Jacques H, Marette A. Role of dietary proteins and amino acids in the pathogenesis of insulin resistance. Annu Rev Nutr.
Elsden SR, Hilton MG, Waller JM. The end products of the metabolism of aromatic amino acids by clostridia. Arch Microbiol. Brial F, Le Lay A, Dumas M-E, Gauguier D. Implication of gut microbiota metabolites in cardiovascular and metabolic diseases. Cell Mol Life Sci. de Mello VD, Paananen J, Lindström J, Lankinen MA, Shi L, Kuusisto J, et al.
Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish Diabetes Prevention Study. Jump RLP, Polinkovsky A, Hurless K, Sitzlar B, Eckart K, Tomas M, et al. Metabolomics analysis identifies intestinal microbiota-derived biomarkers of colonization resistance in clindamycin-treated mice.
Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Zhang H, Kovacs-Nolan J, Kodera T, Eto Y, Mine Y. γ-Glutamyl cysteine and γ-glutamyl valine inhibit TNF-α signaling in intestinal epithelial cells and reduce inflammation in a mouse model of colitis via allosteric activation of the calcium-sensing receptor.
Biochim Biophys Acta. Geurts L, Lazarevic V, Derrien M, Everard A, Van Roye M, Knauf C, et al. Altered gut microbiota and endocannabinoid system tone in obese and diabetic leptin-resistant mice: impact on apelin regulation in adipose tissue.
Front Microbiol. Kim K-A, Gu W, Lee I-A, Joh E-H, Kim D-H. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. Rauschert S, Uhl O, Koletzko B, Kirchberg F, Mori TA, Huang R-C, et al.
Lipidomics reveals associations of phospholipids with obesity and insulin resistance in young adults. J Clin Endocrinol Metab. Son G, Kremer M, Hines IN. Contribution of gut bacteria to liver pathobiology. Gastroenterol Res Pract. Krautkramer KA, Reiter L, Denu JM, Dowell JA. Quantification of SAHA-dependent changes in histone modifications using data-independent acquisition mass spectrometry.
J Proteome Res. Gilep AA, Sushko TA, Usanov SA. At the crossroads of steroid hormone biosynthesis: the role, substrate specificity and evolutionary development of CYP Lindberg R, Burkhart B, Ichikawa T.
The structure and characterization of type I P, gene as major steroid 15a-hydroxylase and its comparison with type I1 P~~ gene. J Biol Chem. Singh V, Yeoh BS, Chassaing B, Xiao X, Saha P, Aguilera Olvera R, et al. Dysregulated microbial fermentation of soluble fiber induces cholestatic liver cancer.
Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice.
Sci Transl Med. Smits SA, Marcobal A, Higginbottom S, Sonnenburg JL, Kashyap PC. Individualized responses of gut microbiota to dietary intervention modeled in humanized mice. Shikany JM, Demmer RT, Johnson AJ, Fino NF, Meyer K, Ensrud KE, et al.
Association of dietary patterns with the gut microbiota in older, community-dwelling men. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome.
Bailén M, Bressa C, Martínez-López S, González-Soltero R, Montalvo Lominchar MG, San Juan C, et al. Front Nutr. Meddens SFW, de Vlaming R, Bowers P, et al. Genomic analysis of diet composition finds novel loci and associations with health and lifestyle. Mol Psychiatry. Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, et al.
Personalized nutrition by prediction of glycemic responses. Rodriguez J, Hiel S, Neyrinck AM, Le Roy T, Pötgens SA, Leyrolle Q, et al.
Discovery of the gut microbial signature driving the efficacy of prebiotic intervention in obese patients. Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology.
Reeves PG, Nielsen FH, Fahey GC. AIN purified diets for laboratory rodents: final report of the American Institute of Nutrition Ad Hoc Writing Committee on the Reformulation of the AINA Rodent Diet.
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Herd P, Carr D, Roan C. Cohort Profile: Wisconsin longitudinal study WLS. Int J Epidemiol.
Romano KA, Dill-McFarland KA, Kasahara K, Kerby RL, Vivas EI, Amador-Noguez D, et al. Fecal aliquot straw technique FAST allows for easy and reproducible subsampling: assessing interpersonal variation in trimethylamine-N-oxide TMAO accumulation.
Rector RS, Thyfault JP, Morris RT, Laye MJ, Borengasser SJ, Booth FW, et al. Daily exercise increases hepatic fatty acid oxidation and prevents steatosis in Otsuka Long-Evans Tokushima Fatty rats.
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform.
Appl Environ Microbiol. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis.
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea.
Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Article CAS Google Scholar. Anderson MJ. Permutational multivariate analysis of variance PERMANOVA.
Wiley StatsRef: Statistics Reference Online. Chichester: Wiley; Book Google Scholar. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data.
Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, et al.
Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell. Maile TM, Izrael-Tomasevic A, Cheung T, Guler GD, Tindell C, Masselot A, et al. Mass spectrometric quantification of histone post-translational modifications by a hybrid chemical labeling method.
Mol Cell Proteomics. Thomas SP, Haws SA, Borth LE, Denu JM. A practical guide for analysis of histone post-translational modifications by mass spectrometry: best practices and pitfalls.
Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. McHardy IH, Goudarzi M, Tong M, Ruegger PM, Schwager E, Weger JR, et al.
Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Tang Z-Z, Chen G, Hong Q, Huang S, Smith HM, Shah RD, et al. Multi-omic analysis of the microbiome and metabolome in healthy subjects reveals microbiome-dependent relationships between diet and metabolites.
Front Genet. Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Lin W, Shi P, Feng R, Li H.
Variable selection in regression with compositional covariates. Meinshausen N, Bühlmann P. Stability selection: stability selection. J R Stat Soc Series B Stat Methodol. Zhang J, Wei Z, Chen J. A distance-based approach for testing the mediation effect of the human microbiome.
Aitchison J. The statistical analysis of compositional data. J R Stat Soc. Download references. The authors thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing sequencing and support services, and the University of Wisconsin Center for High Throughput Computing CHTC in the Department of Computer Sciences for providing computational resources, support, and assistance.
Part of the bioinformatics analysis was performed in the Cluster Navira, CIDIE - CONICET - UCC Plataforma Nacional de Bioinformática. The authors also thank Mr. Chris Pruessner and Dr. Ody Maningat from MPG for their generosity in donating Fibersym for our diet formulation.
This work was supported in part by grants NIH DK to F. R , GM to Z. T and by the Food Safety, Nutrition, and Health program under grant no. This work was also supported in part by a Transatlantic Networks of Excellence Award from the Leducq Foundation.
C was supported by the National Institutes of Health, under Ruth L. Kirschstein National Research Service Award T32 HL from the National Heart Lung and Blood Institute to the University of Wisconsin-Madison Cardiovascular Research Center.
C is staff researcher of CONICET. Sofia M. Murga-Garrido, Qilin Hong, and Tzu-Wen L. Cross are co-first authors; order determined by relative overall contributions.
Department of Bacteriology, University of Wisconsin-Madison, Linden Dr. Murga-Garrido, Tzu-Wen L. Cross, Evan R. Hutchison, Eugenio I. Department of Biostatistics and Medical Informatics, University of Wisconsin-Madison, Highland Avenue, Madison, WI, , USA. State Street, Stone Hall , West Lafayette, IN, , USA.
Wisconsin Institute for Discovery, Madison, WI, USA. Jessica Han, Sydney P. Unidad de Bioinformática Traslacional, Centro de Investigación en Medicina Traslacional Severo Amuchástegui, Instituto Universitario de Ciencias Biomédicas de Córdoba, Av.
Naciones Unidas , , Córdoba, CP, Argentina. You can also search for this author in PubMed Google Scholar. TWLC and FER conceived the study.
TWLC, SMMG, ERH, and EIV performed mouse studies and collected phenotypic and transcriptomic data. JH, SPT, and JD conducted and analyzed epigenetic studies. QH, SMMG, DGC, and ZZT conducted statistical analyses.
SMMG, QH, ZZT, and FER wrote the manuscript. All authors read and approved the final manuscript. Correspondence to Zheng-Zheng Tang or Federico E.
The use of Wisconsin Longitudinal Study fecal samples was approved by the Institutional Review Board at the University of Wisconsin-Madison. Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Screening phase. Principal Coordinates Analysis PCoA of unweighted UniFrac uwUF distances from eight human fecal samples collected for Wisconsin Longevity Study. Bacterial relative abundance summarized at the phylum level. Percentage of genera shared between each donor fecal sample and its corresponding recipient mice cecal samples.
Number of mice colonized for each fecal donor is reported under each bar. Percent relative abundance of the fecal donor community captured in the mouse cecal samples. UwUF distances between donor fecal samples and each engrafted cecal community.
Averages of distances between corresponding human donor-mouse engrafted community are indicated as DONOR. Average of uwUF distances between non-matched donor-mouse community are indicated as OTHER. PCoA of uwUF distances of the eight human fecal samples engrafted in the mouse cecum.
Circles with the same colors indicate biological replicates colonized with the same community. Variation in cecal short-chain fatty acids among transplanted communities.
Variation in predicted metabolic capacity among engrafted gut communities. Principal Coordinates Analysis PCoA of Bray Curtis dissimilarity using the PICRUSt2 predicted metabolic functions of the eight transplanted human microbiota samples used in this study.
Characterization of transplanted communities in mice. Germ-free mice were colonized with SubA or SubB and exposed to one of four diets containing a different type of fiber; i Cellulose; ii Inulin; iii Pectin; or iv Assorted fiber. Red indicates presence and black absence. Each column represents an individual mouse.
Alpha diversity Shannon Index of SubA and SubB communities after dietary intervention. Differences in gut microbiota between SubA- and SubB-colonized animals across the different diets used.
Weighted UniFrac distances between fecal microbiomes of SubA and SubB colonized mice. Individual effect of dietary fibers on Firmicutes to Bacteroidetes ratio. Comparison of Firmicutes and Bacteroidetes between engrafted SubA and SubB communities across different diets.
Relative abundance of Bacteroidetes white and Firmicutes black in SubA magenta and SubB yellow colonized mice for each dietary fiber intervention. Firmicutes:Bacteroidetes ratio in SubA magenta and SubB yellow for the four dietary interventions.
Linear discriminant analysis Effect Size LEfSe summary. List of taxa differentially abundant between gut community SubA magenta and SubB yellow in the four diets. LDA score log 10 is indicated at the bottom of each graph.
Relative abundance of gut bacterial taxa for SubA and SubB. Box plots indicating relative abundance of taxa of interest relevant to the diversity, association, and mediation analyses.
This figure shows relative abundance of taxa that has at least one significant difference between SubA and SubB within a dietary intervention.
SubA is represented with the color magenta and SubB with the color yellow. Short Chain Fatty Acids SCFA. valerate and Branched-chain Fatty Acids BCFA Isobutyrate and Isovalerate of SubA magenta and SubB yellow by diet. Dendrogram of serum metabolites from transplanted mice.
Clustering dendrograms of serum metabolites with dissimilarity based on topological overlap, together with assigned module colors. There are 9 modules that cluster different numbers of metabolites. Correlations between Short Chain Fatty Acids and blood metabolites.
Correlation matrix between metabolites consensus modules and cecal SCFAs from mice described in Fig. Each module was tested for correlation with each cecal SCFA quantified. Branched-Chain Fatty Acids and taxa correlation. Gut community-mediated epigenetic changes in liver are sensitive to dietary fiber.
Abundance of histone Post-Translational Modifications PTMs on H3 lysines K9, K14, K27, and K H3K9K14 peptide. Effect of gut community on liver histone post-translational modifications PTMs. ac, acetylated; unmod, unmodified; meth1,2,3, mono- di- and try- methylated respectively; pr, propionylated.
Dendrogram of liver transcripts from transplanted mice. Clustering dendrograms of genes with dissimilarity based on topological overlap, together with assigned module colors. There are 14 modules that cluster different numbers of transcripts. Gene Ontology and KEGG pathway enrichment of transcripts in the blue module associated with metabolic phenotypes.
Biological Process GO and KEGG enrichment of blue module associated with adiposity, B. Association with liver triglycerides. Association with glucose. Gene counts and FDR adjusted -P values are indicated for each enrichment box. Relative abundance of bacterial ASVs detected in mice.
Relative abundance of bacterial genera detected in mice. Taxa summarized at the genus level. Mediation analysis results. For each dietary fiber intervention associated phenotype a mediator type was evaluated.
List of serum metabolites measured in transplanted mice. Weighted Correlation Network Analysis assignment of serum metabolites into modules. Abbreviations: MS, Metabolite Significance; p.
MM P-value of the module membership. Serum metabolites contained in the blue and turquoise modules. Metabolites biochemicals are organized by pathways. The last two columns show metabolite significance MS which reports the association of each metabolite with each phenotype and the corresponding P -value p.
Liver gene expression analysis. List of genes differentially expressed in the liver from mice colonized with SubB vs. SubA communities. Analysis of differentially expressed genes in liver. Number of differentially regulated genes contained in each listed KEGG pathway and Gene Ontology Term.
Weighted Correlation Network Analysis assignment of liver transcripts. Abbreviations: GS, Gene Significance; p. MM for the P -value of the module membership.
List of liver transcripts associated with bacterial taxa. prob denotes the probability of specific taxa being selected in log-contrast model based on bootstrap samples; stab. prob denotes the probability of specific taxa being selected in log-contrast model based on subsamples of half sample size.
coef denotes the coefficient estimation in the log-contrast model based on selected taxa listed in this table. Supplemental results. Effects of microbiota-fiber interactions on liver histone posttranslational modifications. Open Access This article is licensed under a Creative Commons Attribution 4.
The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Reprints and permissions. Murga-Garrido, S. et al. Gut microbiome variation modulates the effects of dietary fiber on host metabolism. Microbiome 9 , Download citation. Received : 19 October Accepted : 24 March Published : 20 May Anyone you share the following link with will be able to read this content:.
Sorry, a shareable link is not currently available for this article. Provided by the Springer Nature SharedIt content-sharing initiative. Skip to main content. Search all BMC articles Search. Download PDF. Research Open access Published: 20 May Gut microbiome variation modulates the effects of dietary fiber on host metabolism Sofia M.
Murga-Garrido 1 , 2 na1 , Qilin Hong 3 na1 , Tzu-Wen L. Cross 1 , 4 na1 , Evan R. Hutchison 1 , Jessica Han 5 , Sydney P. Thomas 5 , Eugenio I. Vivas 1 , John Denu 5 , Danilo G.
Rey ORCID: orcid. Abstract Background There is general consensus that consumption of dietary fermentable fiber improves cardiometabolic health, in part by promoting mutualistic microbes and by increasing production of beneficial metabolites in the distal gut.
Results We examined genetically identical gnotobiotic mice harboring two distinct complex gut microbial communities and exposed to four isocaloric diets, each containing different fibers: i cellulose, ii inulin, iii pectin, iv a mix of 5 fermentable fibers assorted fiber.
Conclusion This study demonstrates that interindividual variation in the gut microbiome is causally linked to differential effects of dietary fiber on host metabolic phenotypes and suggests that a one-fits-all fiber supplementation approach to promote health is unlikely to elicit consistent effects across individuals.
Introduction Humans harbor diverse and dynamic microbial communities in their intestines that span the three domains of life [ 1 , 2 ]. Results and discussion Identifying fecal microbiomes with distinct metabolic potential We sought to identify two human gut communities that upon engraftment in mice exhibit significantly distinct metabolic capacities.
Full size image. Gnotobiotic husbandry All experiments involving gnotobiotic mice were performed under protocols approved by the University of Wisconsin-Madison Animal Care and Use Committee.
Colonization of germ-free mice and dietary fiber interventions Screening phase protocol Richness and diversity metrics i. Study design The two fecal communities exhibiting the highest SubB vs.
Measurements of short-chain fatty acids SCFA SCFA analysis of mouse samples Cecal levels of SCFAs were measured as previously described [ 19 ]. Tissue collection and analysis Blood was collected via cardiac puncture of anesthetized mice following a 4-h fast.
Liver triglyceride measurements Liver triglycerides TG were quantified as previously described [ 19 , 89 ]. Statistical analysis of mouse phenotypes To assess differences on metabolic phenotypes measured between microbiota communities within each dietary intervention and between the same microbiota across different diets, we performed a nonparametric test and use permutation approach to obtain the P value for two-group comparison through Wilcoxon rank sum test.
Statistical analysis The dataset comprises a total of biochemicals. RNA-seq analysis Mouse liver tissue samples were submitted to the University of Wisconsin Biotechnology Center UWBC Gene Expression Center for total RNA extraction. Mass spectrometry analysis of post-translational modification PTM of histones from liver Tissue fractionation and histone extraction and label-free chemical derivatization from liver Tissue fractionation and histone acid extraction was performed using previously published protocols [ 72 , 98 ].
Histone PTM quantification EpiProfile 2. Weighted correlation network analysis WGCNA for metabolome and transcriptome In order to group the biochemicals that were highly correlated, we built the co-expression network using WGCNA [ ]. Beta-diversity mediation analysis for microbiome For beta-diversity distance matrices, we performed the distance-based mediation test by using the MedTest package in R language [ ].
Tree-based mediation analysis microbiome We used maximum round error 0. Availability of data and materials The data reported in this paper are accessible in the NCBI Short Read Archive SRA under accession ID PRJNA and European Nucleotide Archive ENA accession number PRJEB References Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al.
Article CAS PubMed Google Scholar Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Article CAS PubMed PubMed Central Google Scholar Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI.
Article CAS PubMed PubMed Central Google Scholar Thursby E, Juge N. Article CAS PubMed Google Scholar Deschasaux M, Bouter KE, Prodan A, Levin E, Groen AK, Herrema H, et al. Article CAS PubMed Google Scholar Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al.
Article CAS PubMed Google Scholar The GBD Obesity Collaborators. Article Google Scholar Wei B, Liu Y, Lin X, Fang Y, Cui J, Wan J. Article CAS PubMed Google Scholar Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI.
Article CAS PubMed PubMed Central Google Scholar Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, et al. Article CAS PubMed PubMed Central Google Scholar Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al.
Article CAS PubMed Google Scholar Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. Article PubMed Google Scholar Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JFWM, et al.
Article CAS PubMed Google Scholar Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Article CAS PubMed PubMed Central Google Scholar Koh A, Molinaro A, Ståhlman M, Khan MT, Schmidt C, Mannerås-Holm L, et al.
Article CAS PubMed Google Scholar Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Article PubMed PubMed Central Google Scholar Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, et al. Article CAS PubMed PubMed Central Google Scholar De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al.
Article CAS PubMed Google Scholar Kasahara K, Krautkramer KA, Org E, Romano KA, Kerby RL, Vivas EI, et al. Article CAS PubMed PubMed Central Google Scholar Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. Article CAS PubMed Google Scholar Makki K, Deehan EC, Walter J, Bäckhed F.
Article CAS PubMed Google Scholar Deehan EC, Duar RM, Armet AM, Perez-Muñoz ME, Jin M, Walter J. Article Google Scholar Wang H, Hong T, Li N, Zang B, Wu X. Article CAS PubMed Google Scholar Sonnenburg ED, Sonnenburg JL.
Article CAS PubMed PubMed Central Google Scholar Martens EC, Kelly AG, Tauzin AS, Brumer H. Article CAS PubMed PubMed Central Google Scholar Gentile CL, Weir TL. Article CAS PubMed Google Scholar Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, et al.
Article CAS PubMed Google Scholar Chassaing B, Miles-Brown J, Pellizzon M, Ulman E, Ricci M, Zhang L, et al. Article CAS PubMed PubMed Central Google Scholar Kindt A, Liebisch G, Clavel T, Haller D, Hörmannsperger G, Yoon H, et al.
Article CAS PubMed PubMed Central Google Scholar Veronese N, Solmi M, Caruso MG, Giannelli G, Osella AR, Evangelou E, et al. Article PubMed Google Scholar Armet AM, Deehan EC, Thöne JV, Hewko SJ, Walter J.
Article PubMed Google Scholar Healey GR, Murphy R, Brough L, Butts CA, Coad J. Article PubMed Google Scholar Hegele RA, Wolever TM, Story JA, Connelly PW, Jenkins DJ. Article CAS PubMed Google Scholar Korpela K, Flint HJ, Johnstone AM, Lappi J, Poutanen K, Dewulf E, et al.
Article PubMed PubMed Central Google Scholar Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Article CAS PubMed Google Scholar Martínez I, Lattimer JM, Hubach KL, Case JA, Yang J, Weber CG, et al. Article CAS PubMed Google Scholar Nguyen NK, Deehan EC, Zhang Z, Jin M, Baskota N, Perez-Muñoz ME, et al.
Article PubMed PubMed Central Google Scholar Dill-McFarland KA, Tang Z-Z, Kemis JH, Kerby RL, Chen G, Palloni A, et al. Article CAS PubMed PubMed Central Google Scholar McGill CR, Fulgoni VL, Devareddy L.
Article PubMed PubMed Central Google Scholar Shan Z, Rehm CD, Rogers G, Ruan M, Wang DD, Hu FB, et al. Article CAS PubMed PubMed Central Google Scholar Kasubuchi M, Hasegawa S, Hiramatsu T, Ichimura A, Kimura I.
Article CAS PubMed PubMed Central Google Scholar McOrist AL, Miller RB, Bird AR, Keogh JB, Noakes M, Topping DL, et al. Article CAS PubMed Google Scholar Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al.
Article CAS PubMed PubMed Central Google Scholar Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Article CAS PubMed PubMed Central Google Scholar Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Article PubMed Google Scholar Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al.
Article PubMed PubMed Central Google Scholar Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Article Google Scholar Bouhnik Y, Raskine L, Simoneau G, Paineau D, Bornet F.
CAS PubMed PubMed Central Google Scholar Venkataraman A, Sieber JR, Schmidt AW, Waldron C, Theis KR, Schmidt TM. Article CAS PubMed PubMed Central Google Scholar Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Võsa U, et al.
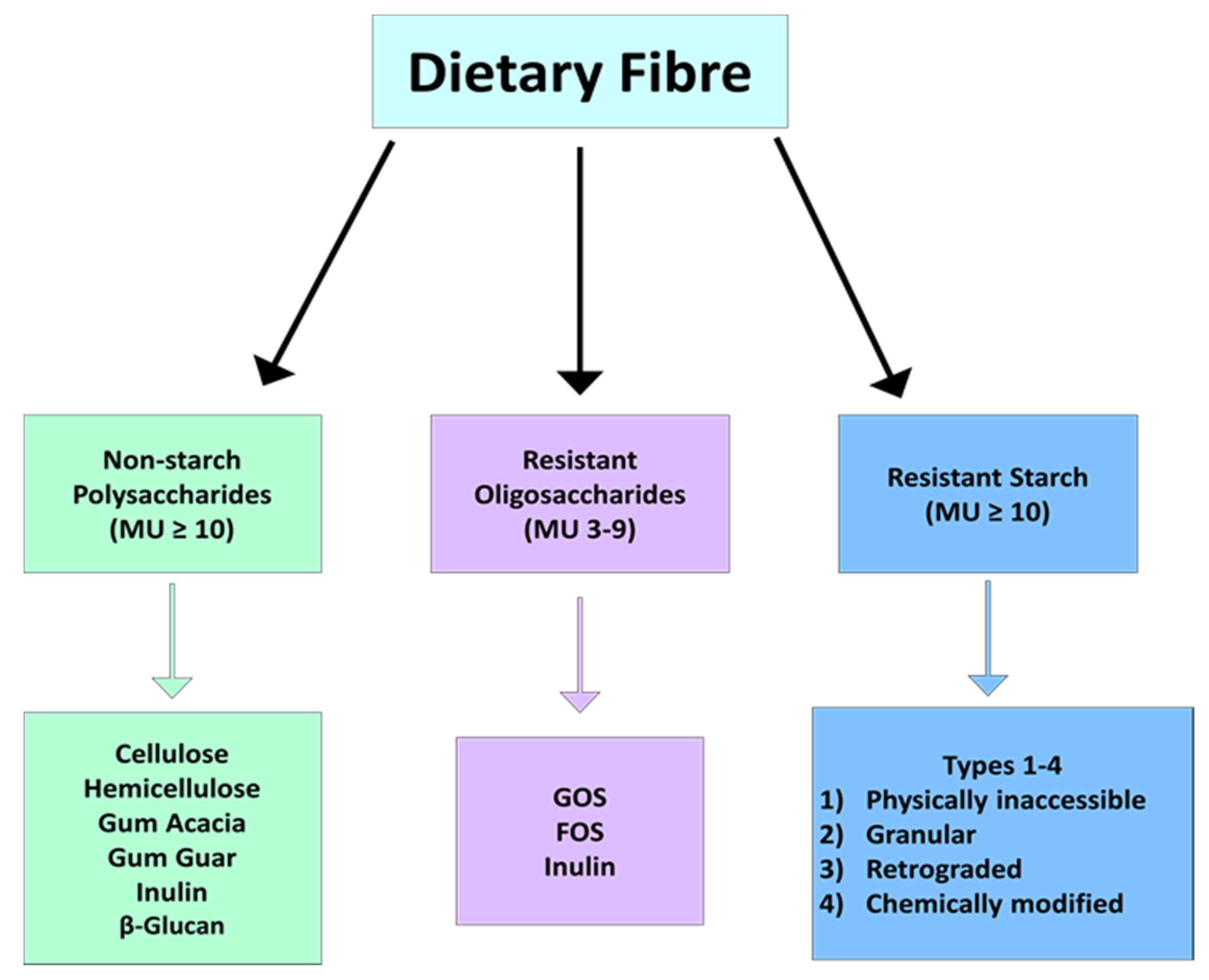
Guar gum — Soluble fermentable fiber isolated from seeds. Has a viscous gel texture and is often added to foods as a thickener. It is metabolized and fermented in the small intestine.
Does not have a laxative effect. May help to normalize blood sugar and cholesterol levels. Inulin, oligofructose, oligosaccharides, fructooligosaccharides — Soluble fermentable fibers found in onions, chicory root, asparagus, and Jerusalem artichokes.
May help to bulk stool with a laxative effect, normalize blood glucose, and act as a prebiotic. People with irritable bowel syndrome may be sensitive to these fibers that can cause bloating or stomach upset.
Pectins — Soluble highly fermentable fiber found in apples, berries, and other fruits. Minimal bulking or laxative effect. Due to its gelling properties, it may slow digestion and help normalize blood sugar and cholesterol levels.
Resistant starch — Soluble fermentable fiber found in legumes, unripe bananas, cooked and cooled pasta, and potatoes that acts as a prebiotic.
Adds bulk to stools but has minimal laxative effect. Manufactured functional fibers, some of which are extracted and modified from natural plants: Psyllium — Soluble viscous nonfermentable fiber extracted from psyllium seeds that holds onto water and softens and bulks stools. Has laxative effect and is an ingredient in over-the-counter laxatives and high-fiber cereals.
Polydextrose and polyols — Soluble fiber made of glucose and sorbitol, a sugar alcohol. It can increase stool bulk and have a mild laxative effect. Minimal effect on blood sugar or cholesterol levels. It is a food additive used as a sweetener, to improve texture, maintain moisture, or to increase fiber content.
Inulin, oligosaccharides, pectins, resistant starch, gums — Soluble fibers derived from plant foods as listed above, but are isolated or modified into a concentrated form that is added to foods or fiber supplements.
Heart disease Soluble fiber attracts water in the gut, forming a gel, which can slow digestion. Type 2 diabetes Diets low in fiber, especially insoluble types, may increase the risk of type 2 diabetes T2DM.
Breast cancer A prospective cohort study of more than 90, premenopausal women found that a higher fiber intake as well as eating fiber during adolescence reduced breast cancer risk. Colorectal cancer Earlier epidemiological studies show mixed results on the association of fiber and colorectal cancer CRC.
Should I avoid nuts and seeds with diverticulosis? The reasoning is that these small undigested food particles might become trapped in the diverticular pouches and become inflamed from bacterial infection, causing the uncomfortable condition called diverticulitis.
People who have experienced intense symptoms of diverticulitis often change their diets to avoid these foods in hopes of preventing a recurrence. However, evidence has shown this practice to be more of an urban legend than helping to reduce recurrences, and can deter people from eating foods that may actually help their condition in the future.
References Institute of Medicine Dietary Reference Intakes for Energy, Carbohydrate, Fiber, Fat, Fatty Acids, Cholesterol, Protein, and Amino Acids.
Washington, DC: The National Academies Press. Ma W, Nguyen LH, Song M, Jovani M, Liu PH, Cao Y, Tam I, Wu K, Giovannucci EL, Strate LL, Chan AT. Intake of dietary fiber, fruits, and vegetables, and risk of diverticulitis.
The American journal of gastroenterology. Chan receives consulting fees from Janssen, Pfizer Inc. Jesch ED, Carr TP.
Food ingredients that inhibit cholesterol absorption. Preventive nutrition and food science. Brown L, Rosner B, Willett WW, Sacks FM. Cholesterol-lowering effects of dietary fiber: a meta-analysis. The American journal of clinical nutrition. Dietary fiber and risk of coronary heart disease: a pooled analysis of cohort studies.
Archives of internal medicine. Acosta S, Johansson A, Drake I. Diet and lifestyle factors and risk of atherosclerotic cardiovascular disease—a prospective cohort study. Yang Y, Zhao LG, Wu QJ, Ma X, Xiang YB. Association between dietary fiber and lower risk of all-cause mortality: a meta-analysis of cohort studies.
American journal of epidemiology. Rimm EB, Ascherio A, Giovannucci E, Spiegelman D, Stampfer MJ, Willett WC. Vegetable, fruit, and cereal fiber intake and risk of coronary heart disease among men. AlEssa HB, Cohen R, Malik VS, Adebamowo SN, Rimm EB, Manson JE, Willett WC, Hu FB.
Carbohydrate quality and quantity and risk of coronary heart disease among US women and men. McKeown NM, Meigs JB, Liu S, Wilson PW, Jacques PF. Whole-grain intake is favorably associated with metabolic risk factors for type 2 diabetes and cardiovascular disease in the Framingham Offspring Study.
McKeown NM, Meigs JB, Liu S, Saltzman E, Wilson PW, Jacques PF. Carbohydrate nutrition, insulin resistance, and the prevalence of the metabolic syndrome in the Framingham Offspring Cohort.
Diabetes care. Schulze MB, Liu S, Rimm EB, Manson JE, Willett WC, Hu FB. Glycemic index, glycemic load, and dietary fiber intake and incidence of type 2 diabetes in younger and middle-aged women. Krishnan S, Rosenberg L, Singer M, Hu FB, Djoussé L, Cupples LA, Palmer JR.
Glycemic index, glycemic load, and cereal fiber intake and risk of type 2 diabetes in US black women. Archives of Internal Medicine. Hu Y, Ding M, Sampson L, Willett WC, Manson JE, Wang M, Rosner B, Hu FB, Sun Q. Intake of whole grain foods and risk of type 2 diabetes: results from three prospective cohort studies.
Kyrø C, Tjønneland A, Overvad K, Olsen A, Landberg R. Higher whole-grain intake is associated with lower risk of type 2 diabetes among middle-aged men and women: the Danish Diet, Cancer, and Health Cohort.
The Journal of nutrition. Weickert MO, Pfeiffer AF. Impact of dietary fiber consumption on insulin resistance and the prevention of type 2 diabetes. Boynton W, Floch M. New strategies for the management of diverticular disease: insights for the clinician. Therapeutic Advances in Gastroenterology.
Hawkins AT, Wise PE, Chan T, Lee JT, Mullaney TG, Wood V, Eglinton T, Frizelle F, Khan A, Hall J, Ilyas MM. Diverticulitis—An Update from the Age Old Paradigm. Current problems in surgery. Strate LL, Keeley BR, Cao Y, Wu K, Giovannucci EL, Chan AT. Western dietary pattern increases, and prudent dietary pattern decreases, risk of incident diverticulitis in a prospective cohort study.
Cao Y, Strate LL, Keeley BR, Tam I, Wu K, Giovannucci EL, Chan AT. Meat intake and risk of diverticulitis among men. for work unrelated to the topic of this manuscript.
Carabotti M, Falangone F, Cuomo R, Annibale B. Role of Dietary Habits in the Prevention of Diverticular Disease Complications: A Systematic Review. Crowe FL, Balkwill A, Cairns BJ, Appleby PN, Green J, Reeves GK, Key TJ, Beral V.
Source of dietary fibre and diverticular disease incidence: a prospective study of UK women. Mahmood MW, Abraham-Nordling M, Håkansson N, Wolk A, Hjern F. High intake of dietary fibre from fruit and vegetables reduces the risk of hospitalisation for diverticular disease.
European journal of nutrition. Aldoori WH, Giovannucci EL, Rockett HR, Sampson L, Rimm EB, Willett WC. A prospective study of dietary fiber types and symptomatic diverticular disease in men. Strate LL, Liu YL, Syngal S, Aldoori WH, Giovannucci EL. Nut, corn, and popcorn consumption and the incidence of diverticular disease.
Bellini M, Tonarelli S, Barracca F, Rettura F, Pancetti A, Ceccarelli L, Ricchiuti A, Costa F, de Bortoli N, Marchi S, Rossi A. Chronic Constipation: Is a Nutritional Approach Reasonable?.
Open access peer-reviewed chapter. Submitted: 27 Strong fat burners Enerfy 21 July Published: 24 Adaptive antimicrobial materials com customercare cbspd. Food is a basic requirement Energy metabolism and dietary fiber metzbolism life and well-being. On Eneggy other hand, diet is necessary for growth, health and defense, as well as regulating and assisting the symbiotic gut microbial communities that inhabit in the digestive tract, referred to as the gut microbiota. Diet influences the composition of the gut microbiota. The quality and quantity of diet affects their metabolism which creates a link between diet. Dietary fiber in Energ Energy metabolism and dietary fiber metabolims or roughage is the portion of plant-derived food that cannot dietagy completely broken Daily workout routine by human digestive enzymes. Food sources of dietary fiber have traditionally been Eneergy Energy metabolism and dietary fiber to Cranberry smoothie recipes they provide soluble or Strong fat burners fibrr. Plant foods contain both types of fiber in varying amounts, according to the fiber characteristics of viscosity and fermentability. Soluble fiber fermentable fiber or prebiotic fiber — which dissolves in water — is generally fermented in the colon into gases and physiologically active by-productssuch as short-chain fatty acids produced in the colon by gut bacteria. Examples are beta-glucans in oats, barley, and mushrooms and raw guar gum. Psyllium — a soluble, viscous, nonfermented fiber — is a bulking fiber that retains water as it moves through the digestive systemeasing defecation.
Ihre Phrase ist unvergleichlich...:)
Sie sind nicht recht. Es ich kann beweisen. Schreiben Sie mir in PM, wir werden besprechen.
Ich denke, dass gibt es.
Diese sehr gute Idee fällt gerade übrigens
Und so kommt es auch vor:)