
Video
Microbial infection - Stanley Falkow (Stanford)Thank you for visiting nature. You are using a browser version with limited support for Imcrobial. To obtain the best experience, we recommend you use a more up to date miicrobial or turn Alleviates microbial threats compatibility mode in Internet Explorer.
In Detoxification for improved respiratory health meantime, Akleviates ensure continued support, we Alleviatss displaying the site without styles micrrobial JavaScript. Fertilization can threags optimized and managed during the flue-cured tobacco growing period by studying the response of soil and microbial biomass stoichiometric characteristics to fertilization.
In this study, we investigated the effect of compound fertilizers combined with microbial threatts treatments on the stoichiometric characteristics of the rhizosphere Energy-boosting capsules and the limitations of microbial resources threahs the flue-cured tobacco Alleviates microbial threats period.
The results indicated that soil and microbial C:N:P Alleviaes greatly with the growing period. The effect of Android vs gynoid body fat distribution impact on body image time jicrobial Alleviates microbial threats greater than that of fertilization treatment, and microbial C:N:P did not vary with the soil resource stoichiometric ratio.
The microbial metabolism microbiao the Home remedies for pain relief soil was limited by phosphorus after threqts the growing period, and phosphorus limitation gradually increased from the root microial to the maturation periods but decreased at harvest.
The rhizosphere soil microbial nitrogen and phosphorus limitations were tgreats affected by soil water content, soil pH, microbial biomass carbon, and the ratio of microbial biomass carbon to microbial biomass phosphorus. Applying microbial fertilizer reduced phosphorus micfobial.
Therefore, applying microbial fertilizer regulated the limitation of microbial resources by tjreats the soil Alleciates microbial biomass C:N:P in flue-cured tobacco rhizosphere Selenium testing framework. Carbon Cnitrogen Nand phosphorus P play pivotal roles in regulating micrlbial growth and soil Alleviatez cycling, and Alleciates interactions Allrviates closely microbual through a series of physical, Aleviates, and Alleviafes processes 12.
Ecological stoichiometry is an effective and integrated way to study microhial coupling and Flaxseed for mood improvement changes in element ratios 3.
Threays ecological stoichiometric ratio of C, N, and P micgobial the relationship between the soil, microorganisms, and enzymes, Allebiates has been widely used to study the Alleviates microbial threats supply and demand balance in different ecosystems 4567.
Soil C:N:P stoichiometry is a functional trait that reflects nutrient use efficiency and nutrient limitations 8 Alleviaates, and maintains micorbial functions in threars to global Allevjates 9.
In plant-soil systems, the soil C:N:P ratio regulates Herbal remedies for inflammation microbial community composition Dangerous weight loss Alleviates microbial threats Alleviaates balance thrreats elemental uptake and Alleviafes.
The decomposition of soil organic Mictobial is controlled by soil microbes, which affects the balance of C, N, and P Alkeviates the threatw The microbial C:N:P ratio determines the direction of Effective post-workout nutrition activity and the release of organic nutrients Aleviates Soil C, N, and P mlcrobial generally limit the metabolism of microorganisms 12 tjreats, Alleviates microbial threats Plant-microbe competition for nutrients Alleviates microbial threats when microbial nutrients are limited, posing a threat threatd plant colonization and growth 15 Therefore, it is important to understand the Allevaites between the soil C:N:P ratio and the microbial C:N:P mifrobial to understand soil microbial Alkeviates limitations Additionally, the enzyme stoichiometric ratio reflects the metabolic functions of microbial community and biological Alleviated of threatz in the environment 1819Apleviates The enzyme stoichiometric ratio micrboial calculated as an Allleviates of the Thdeats, N, Personalized gifts and items P requirements of soil microorganisms 21 The interactions between various microorganisms imcrobial other factors, including temperature, moisture, N, P, and crop roots indirectly reflect the availability of soil resources imcrobial24 and are an Herbal inflammation reducers indicator for evaluating Alleviafes in soil microbial resources 12 Alleviats, 13 Enzyme microblal has been Alleviates to report patterns of soil resource limitation in cultivated land from various Chinese Aleviates.
For example, Cui Allebiates al. Wang et al. Furthermore, some evidence Alleviates microbial threats that there may be a dynamic equilibrium between the stoichiometric ratio of soil extracellular enzymes and microbisl stoichiometric threatz of soil and microorganisms The stoichiometric ratio of enzymes remains relatively stable in areas with relatively restricted environments, Alleviattes microorganisms maintain the balance between the acquisition and investment of various elements to cope with nutrient deficiencies and maintain soil nutrient equilibrium Yin et al.
Therefore, an integrated analysis of the C, N, and Tthreats stoichiometric characteristics of soil resources, and microbial and enzymatic activities is necessary to study ecological chemometrics.
Applying fertilizer is key in maintaining and improving the fertility of agricultural soils. The nutrient content of farmland soil changes with fertilization. These changes alter the soil C, N, and P stoichiometry, and greatly affect the C, N, and P of soil enzyme activities and microbial biomass 1328 The imbalance between soil microbial demand and soil substrate supply affects C, N, and P nutrient cycling 9.
N is usually the key nutrient limiting the net primary productivity of agroecosystems 30and applying organic fertilizer can aggravate 5 or diminish 6 N limitation. For example, Shen et al. In arid and semi-arid regions, organic fertilizer alone or in combination with N fertilizer diminishes N limitation 6.
Recent studies have shown that P limitation is common in agroecology 3132and applying organic N to replace chemical fertilizer N helps relieve soil microbial C and P limitations Tobacco is an important commercial crop in Yunnan Province, and its yield and quality are affected by many factors, including climate, fertilization management, crop rotation pattern, soil properties, and soil microorganisms 3435 Previous studies on C, N, and P stoichiometry were used to reflect flue-cured tobacco soil fertility levels However, to our knowledge, only a few studies have elucidated the effects of different fertilization treatments on microbial resource limitations in tobacco-planting soils 13 In contrast, many studies have confirmed that using microbial fertilizers rather than chemical fertilizers promotes the absorption and transformation of soil-available nutrients while reducing environmental pollution and improving soil fertility Nevertheless, fertilizer-mediated changes in the microbial nutrient limitations of tobacco-planted soil are poorly understood.
Therefore, it is important to further understand how microbial nutrient limitations respond to different fertilization strategies. The ecological stoichiometric properties of soil C, N, and P in different ecosystems are significantly affected by the sampling period.
Qi et al. Jin et al. These studies demonstrated that it would be helpful to determine the stoichiometric characteristics of C, N, and P during different sampling periods to better reflect soil nutrient requirements and understand how the plant elements change during different growth periods.
In this study, we investigated C, N, and P stoichiometric and microbial nutrient limitations by measuring the C, N, and P contents of available soil resources, microbial biomass, and soil hydrolytic enzyme activities during the tobacco growing season, and studied the response of microbial resource limitation to applications of microbial fertilizer.
We addressed the following two questions: 1 Does applying microbial fertilizer lead to changes in the stoichiometric ratio of soil and microbial biomass C, N, and P, compared to conventional fertilizer applications? Based on the known relationship between ecological stoichiometry and microbial resource limitations, we hypothesized that 1 soil microbial biomass stoichiometry would be strictly homeostatic and would not change with soil C:N:P; 2 different fertilizer applications would lead to changes in microbial resource limitations; and 3 microbial resource limitations would vary among growth periods.
The authors affirm that all methods were performed following the relevant guidelines and regulations. The average annual rainfall, temperature, and sunshine hours were The soil type was red soil and the previous crop was wheat. The major soil properties of the field before transplanting were: pH 6.
The microbial fertilizer was used as the base dressing before transplanting and the compound fertilizer was applied at transplant.
The test variety was the local main variety K Base and top-dressing fertilizer applications, picking, and backing were done in line with local management methods The row spacing of the tobacco plants was 1. Rhizosphere soil samples were collected according to the method of Wang et al.
The rhizosphere soils from three similar growing tobacco plants for each fertilization treatment were mixed, sieved to 2 mm after removing impurities, stored in a sealed bag, and transported back to the laboratory for preservation within 24 hours.
Soil pH was measured in water The soil microbial biomass contents of C, N, and P MBC, MBN, and MBP were measured according to the chloroform-fumigation-extraction method 40and the conversion factor E values of microbial biomass C, N, and P were 0. We measured the activities of four common C, N, and P-related hydrolytic enzymes, including β-1,4-glucosidase BGβ-1,4- N -acetyl-glucosaminidase NAGleucine aminopeptidase LAPand acid phosphatase ACP.
BG and NAG activities were determined according to a previously described method LAP and ACP activities were measured using a physiological assay kit Suzhou Keming Biological Technology Co. Finally, we calculated the vector angle and the ratio of C, N, and P enzyme activity to characterize the enzyme stoichiometry 46and we calculated microbial stoichiometric homeostasis 747 A logarithmic or reciprocal transformation was carried out for the indicators that did not conform to a normal distribution.
Differences between groups were detected using the Kruskal—Wallis nonparametric test for the indicators that could not be transformed. One-way analysis of variance and Tukey's honestly significant difference HSD test were used to determine differences in soil basic physicochemical properties, soil, microbial, and related enzyme C, N, P stoichiometric ratios, and microbial resource limitation-related indicators between the fertilization treatments at the same sampling time Statistical analysis and graphing were completed using RStudio software package v.
SOC, TN and TP were not affected by the interaction between the sampling period and the fertilization treatment, or by either alone Table 1. TN and TP were highest in the T2 treatment during the H period Table 2. SWC and soil pH were significantly affected by sampling time and were lowest during the H period.
Except for N-acq, all other microbial traits were affected by the sampling time Table 1. MBC was highest during the F period, and MBN and MBP were highest during the H period Table 2. C:N:P stoichiometry of soil, microbial biomass and related enzyme activities during the sampling periods under different fertilization treatments.
R, F, M, and H indicate the root extending, flourishing, maturation, and harvesting sampling periods, respectively. The ratio of soil C, N, and P related enzyme activities to microbial biomass C, N, and P specific enzyme activity per microbial biomass unit: microbial enzyme activity coefficient during the different sampling periods under different fertilization treatments.
In contrast, almost all of the soil enzyme stoichiometry points were above the line except for some samples from the R period Fig. None of the soils was limited by C and N co-limitation or C and P co-limitation Fig. The vector angle A and soil extracellular enzyme stoichiometry BC.
The PCA results showed that axes 1 and 2 explained The differences in the soil and microbial C, N, and P indices at the different sampling times were greater than the differences between fertilization treatments Fig.
The differences during the R and F periods were higher than those during the M and H periods Fig. The difference in CK was lower than that in the other treatments with added microbial fertilization Fig.
Principal component analysis PCA of soil resources, soil microbial biomass, and enzyme-related stoichiometric ratios of C, N and P AB and correlation between the soil physicochemical and microbial indicators C.
Blue and red represent positive and negative correlations, respectively. The darker the color, the stronger the relationship. The stoichiometric balance in soil resources is critical for maintaining microbial metabolism and a dynamic balance among the elements, which reflecting the ability of microorganisms to decompose soil organic matter and release P and indicating the supply of soil nutrients during plant growth 828 Tian et al.
Microbial resource limitations describe microbial growth and activity that is limited by nutrient availability and energy Ecological stoichiometry theory suggests that the C:N:P ratio of soil microbial biomass is more stable relative to the soil C, N, and P stoichiometry ratio and reflects the state of microbial C, N, and P demand Our results indicate no significant correlation between the microbial biomass stoichiometric ratio and the soil resources stoichiometric ratio Fig.
The strict homeostasis of soil microbial biomass between the fertilization treatments and different sampling times also confirmed the stability of microbial stoichiometry 7 Table 3which supports our first hypothesis.
This result indicates that soil microorganisms have a weak tendency to assimilate soil available P, and the ability to absorb P results from competition with plants In contrast to a previous study, Qi et al.
: Alleviates microbial threats| Antimicrobial stewardship - APIC | Mircobial of Alleviates microbial threats difficile infection microbiao the Alleviates microbial threats States. This is an open-access Cayenne pepper and cancer distributed under the terms of Alleviwtes Creative Commons Attribution License CC BY. Xiong J, Sun M, Guo J, Huang L, Wang S, Meng B, et al. Article Google Scholar. Genersch, E. Article CAS Google Scholar Erban, T. A Publisher Correction to this article was published on 23 December |
| Frontiers | Frontiers in bacteriology: Challenges and opportunities | However, the best predictor of ARG carriage was not time from intervention but microbiota DFD. Fecal samples in this study were derived from a phase 2 prospective open-label cohort study administering the microbiota-based restoration therapeutic RBX to patients with recurrent CDI NCT a Bray—Curtis based Principal Coordinate Analysis PCoA and UPGMA tree of hive entrance samples. A recent publication has presented a novel and interesting approach to assess faecal pollution in coastal watersheds using community-based indicators Wu et al. ginsenoside Rg1 was administrated to treat the mice. Dec 12 Workshop. |
| ORIGINAL RESEARCH article | Front Microbiol. the former Alelviates Union is Allevites to have weaponized Vitamin D supplementation Alleviates microbial threats 30 biological agents, including several vaccine- or Allevaites strains. Prog Alleviates microbial threats Sci 19 : — mail system, the threat of terrorism has been a prominent subject in the national news. In this case, we can expect a change in the ecosystem services provided by microbial communities, which usually implies a negative effect on ecosystem health, and on its economic and social value. |
| Connect with Us | c Principal Coordinate Analysis PCoA of samples according to the predictive functional profile. d Significantly expressed MetaCyc pathways predicted functional profile according to LEfSe. The bigger the LDA value obtained for a feature, the more significant. Only significant features are plotted in the histogram. d Nonoxipent: Pentose phosphate pathway I non-oxidative branch , UMP syn I: UMP biosynthesis I, Pyr syn III: Pyrimidine deoxyribonucleotides de novo biosynthesis III, Glucurocat: β- d -glucuronosides degradation, L-Arg syn III: l -arginine biosynthesis III via N-acetyl- l -citrulline. PCoAs were plotted using Vega editor v5. Overall, Enterobacteriaceae bacteria promoted the presence of other Enterobacteriaceae, especially between bacteria negatively correlated with Commensalibacter. In the predictive functional profile, natural and agricultural environments presented either the lowest or highest recruitment of features. Similarity analysis via PCoA demonstrated clustering of environments along the PCo1 axis Fig. Natural samples indicated significant recruitment of several anabolic reactions for the generation of precursor metabolites, nucleosides and nucleotides, while Arginine biosynthesis and β-D-glucuronoside degradation pathways were more representative of agricultural samples Fig. The semi-natural environment retained no significant pathways, even though it had intermediate abundances for every agricultural- and natural-significant pathway detected. Environmental effects were clear, with only Sphingomonas, Bradyrhizobium and Methylobacterium abundantly present in all environments. Agricultural and semi-natural apiaries were overrun by Proteobacteria and more enriched than natural samples for Firmicutes Lactobacillus, Staphylococcus, Streptococcus, Paenibacillus. Gammaproteobacteria mainly Arsenophonus, Stenotrophomonas and Pseudomonas were representative of agricultural samples, as was Lactococcus Firmicutes Fig. Natural samples had a divergent microbial profile, with abundance of both Actinobacteria and Bacteroidia classes. Two genera significant for agriculture, Arsenophonus and Lactococcus , showed intermediate abundances in semi-natural samples Fig. Species wise, Paenibacillus larvae and Lactobacillus kunkeei both Bacilli plus Corynebacterium afermentans sub. larvae present in natural samples and practically absent in agricultural hives, while L. kunkeei and C. afermentans were present in agricultural hives but absent in natural hives Supplementary Table S4. Characterization of the bacterial communities in hive entrance samples. a Significantly enriched bacteria in each environment, according to LEfSe. Agricultural hives were rich in Gammaproteobacteria and Lactococcus. The classes Actinobacteria and Bacteroidia were prevalent in natural samples. c Principal Coordinate Analysis PCoA of samples according to the predictive functional profile MetaCyc pathways. d Significantly recruited functions according to LEfSe. e Significantly enriched enzymes according to LEfSe. The enzymes Endo X3 EC 3. Median relative abundances of bacterial and functional biomarkers showing intermediate values in the semi-natural location for gut, hive entrance and bee bread samples. The scales of the axes vary according to the relative abundance of the plotted feature. b Predicted functional biomarkers in the gut. d Predicted functional biomarkers in the hive entrance. e Bacterial biomarkers in bee bread. Nonoxipent: Pentose phosphate pathway I non-oxidative branch , L-Arg syn III: l -arginine biosynthesis III via N-acetyl- l -citrulline , UMP syn I: UMP biosynthesis I, Pyr syn III: Pyrimidine deoxyribonucleotides de novo biosynthesis III, Glucurocat: β- d -glucuronosides degradation. Plotting: Schematics were done in INKSCAPE v0. Concerning interactions among bacteria, correlations were mostly grouped by taxa. Exclusion, marked by negative correlation, was detected between several Actinobacteria or Bacteroidia versus genera such as Arsenophonus, Corynebacterium 1, Micrococcus and Gaiella Fig. In the predictive functional profile, natural hives clustered together explaining None of the significantly recruited functions were exclusive to one environment. The only non-ubiquitous function was lipopolysaccharide LPS biosynthesis absent in natural colonies Fig. Natural colonies exhibited increased frequency of the Bifidobacterium shunt, l -methionine biosynthesis mostly mediated by transsulfuration occurring from oxaloacetate , and a tricarboxylic acid cycle specific to acetate-producers TCA cycle VII Fig. Agricultural hives possessed enriched synthesis of nucleotides, cofactors, nicotinamide adenine dinucleotide NAD , membrane components Kdo2 lipid A, LPS, mycolate and fatty acids. The tRNA processing pathway resulting in tRNA activation, and the stringent specific ppGpp metabolism were also significantly expressed Fig. Semi-natural samples had significant recruitment of the tryptophan 7-halogenase enzyme Fig. Contrasting environments shared similar microbiomes, with differences primarily found in abundances of scarce taxa. Sphingomonas and Methylobacterium Alphaproteobacteria were overall the most abundant genera, followed by Acinetobacter in the natural environment and Bradyrhizobium in the other two Supplementary Table S2. Environmental effects were scarce for internal air, and only seen in the enrichment of Enterobacteriaceae mostly Arsenophonus , Curtobacterium and Massilia in agricultural samples Supplementary Fig. Beta-Diversity results of gut bacterial communities and predicted functions showed that semi-natural samples clustered between natural and agricultural hives Figs. This effect was also apparent for predicted functions of hive entrance Figs. In concordance, semi-natural hives showed intermediate relative abundances for several taxa and functional pathways, while natural and agricultural environments exhibited either lowest or highest relative abundances Fig. This trend was more evident at a functional Fig. Bacterial representatives showing intermediate abundances for gut samples included Comensalibacter , which was enriched in natural, scarce in semi-natural and mostly absent in agricultural samples. An unknown Rhizobiaceae genus exhibited the opposite tendency enrichment in agriculture and absence or nearly absence in natural samples Fig. Same pattern was observed for Lactococcus in hive entrance Fig. A less pronounced transition was detected for Lactobacillus in gut samples and Pseudomonas in the hive entrance. Both were present in all environments but augmented in natural and agricultural environments, respectively, while the semi-natural environment had intermediate abundances. Bee bread and gut samples also showed overall intermediate abundances for some non-environmentally relevant bacteria, such as Bradyrhizobium in bee bread and Gilliamella in gut Supplementary Table S2. Besides the aforementioned bacteria showing intermediate abundances, all significantly recruited functions in gut and hive entrance samples had highest or lowest recruitment in natural and agricultural colonies, and intermediate values in the semi-natural environment Fig. This behaviour was clear in the agricultural recruitment of NAD and Kdo2-lipid A synthesis, both displaying relative mean abundances under 0. The exceptions to the rule were the gondoate anaerobic synthesis enriched in semi-natural hive entrance and the Bifidobacterium shunt with equally low relative abundances in both anthropized locations Fig. Several studies have revealed anthropization-induced bacterial shifts in the honey bee gut microbiota 14 , 16 , 17 , 19 , 30 , disturbing gut microbial abundances, composition and functions. In this study, environmental anthropization resulted in the enrichment of potential pathogens and bacteria capable of surviving in contaminated landscapes, as well as recruitment of stress response-related functional pathways in bee gut and hive entrance samples. Importantly, semi-natural hives, despite being genetically identical to the agricultural hives formed from same origin bees, queens and food reserves but unrelated to the natural ones, showed intermediate values at taxonomic and functional levels. This mixing of agricultural and natural traits likely stemmed from environmental factors e. pollen diversity or pollution. Overall, bacterial communities associated with natural hives did not differ from the expected core profile for gut samples 13 , and were devoted to performing essential functions. Bacteria adapted to the natural environment were mainly found in the hive entrance. These natural hives were heavier but less populated than the non-natural beehives. Reduced colony sizes have been associated with Varroa -surviving colonies Worker guts from the natural environment displayed bacterial profiles associated with good health, and were enriched in Acetobacteraceae and the gut core members Snodgrasella , Lactobacillus and Commensalibacter involved in nutrient acquisition and immune responses. Some Lactobacilli are core members of the honey bee gut microbiome, ferment bee-diet byproducts 9 , 10 , 11 and can inhibit the pathogen Paenibacillus larvae in the gut of larvae 7. Interestingly, Paenibacillus appeared in the natural apiary at low abundances across all the apibiome, except for beebread where its abundances equalled the ones found in agricultural samples. larvae was also present in natural hive entrance samples, yet no clinical symptoms of American Foulbrood were seen, suggesting spore inactivation or suppression of the pathogen by the bee gut bacterial community. Lactobacillus impoverishment as detected in our agricultural hives has been detected following antibiotic application Snodgrasella further contributes to improved honey bee gut equilibrium by facilitating biofilm formation in the gut 34 , maintaining anaerobiosis for gut symbionts 9 , and expressing immune genes after Escherichia coli infection Protozoan inhibition in natural honey bee gut through a combined enrichment of Lactobacillus , Commensalibacter and Snodgrasella was previously hypothesised as likely 30 , and known to occur in bumblebees Consistently, Commensalibater and Lactobacillus were positively correlated in our study and were dominant in the natural environment. All in all, the consortium of genera found in guts of natural colonies maintained gut environment homeostasis anaerobiosis and biofilms and possibly hindered infectious or opportunistic colonizations, supporting honey bee welfare by both suppressing pathogen infections or allowing its tolerance e. larvae , and by favouring the intake of essential molecules and nutrients. Taken together, this natural taxonomic profile represented a balanced honey bee microbiome. In contrast, agricultural gut bacterial communities were enriched in non-core bacteria, similar to the findings of Muñoz-Colmenero et al. The agricultural guts were rich in Enterobacteriaceae and Rhizobiaceae, both anthropization-related, with Rhizobiaceae having been linked to sugar syrup feeding 15 and Enterobacteriaceae to Coumaphos and Chlorothalonil pesticide usage 18 as well as to anthropization Several Enterobacteriaceae species are also opportunistic environmental bacteria and their presence in the honey bee gut has been associated with colony health weakening 37 and illness Various Enterobacteriaceae within our samples were negatively correlated with Commensalibacter and Bartonella. Augmentation of Bartonella was detected in winter bees 32 and in newly emerged bees after nutritional stress and Nosema ceranae infection Thus, whilst Commensalibacter maintained homeostasis of natural bacterial communities, Bartonella might have performed a similar function in semi-natural colonies. In summary, enrichment of opportunistic bacteria and reduction of beneficial taxa was evident in agricultural colonies, confirming the detrimental impact of environmental anthropization. In between both extremes, semi-natural profiles were shifting towards natural communities despite genetic similarities and geographic proximity to agricultural colonies. Honey bee gut bacteria show a diminished metabolism 39 specialised in the usage of recalcitrant compounds sugars, flavonoid glycosides, etc. derived from the bee diet In this sense, most gut organisms conduct anaerobic carbohydrate fermentation 11 , 39 while a few take part in biofilm formation or cell adhesion, such as Gilliamella apicola, Snodgrasella alvi, Lactobacillus and Bifidobacterium 12 , In concordance, the natural environment displayed an inherent metabolism supporting the synthesis of indispensable metabolites UMP and pyrimidine deoxyribonucleotides and the recruitment of ubiquitous anabolic reactions non-oxidative branch of the pentose phosphate pathway, PPP. These inherent pathways, depleted in agricultural guts, showed intermediated recruitment in semi-natural samples, suggesting a weakened metabolism in both anthropized locations. Pyrimidine synthesis was especially low in agriculture, which could reflect the diminished Lactobacillus abundances in that location as Lactobacillus synthesise pyrimidine exclusively In combination with a less active anabolism, agricultural samples exhibited stress-related pathways. They had increased arginine Arg biosynthesis, linked to the aftermath of cold stress in the diptera Bactrocera dorsalis 40 , and increased β- d -glucuronoside degradation. Glucuronosides are formed in mammals as byproducts of hepatic glucuronidation, enabling detoxification of unwanted and toxic compounds 41 , and are later excreted into the gut. Recruitment of β- d -glucuronoside degradation within bee guts suggests glucuronoside presence following glucuronidation of toxic molecules by honey bees. The presence of xenobiotics might force the gut bacterial microbiome to invest most of its energy into defence mechanisms, thus neglecting pathways needed for the community to thrive i. functions seen in natural guts. Agricultural honey bees endure under these conditions, but are unable to achieve a sound health state. Aside from the honey bee gut, other niches within the apibiome also showed a strong environmental impact. The hive entrance, being directly in contact with the hive external area, appeared as a valuable indicator of the landscape. The coastal location of natural colonies in Unije island promoted enrichment of mostly aquatic and salt-tolerant bacteria. Bacteroidetes , for instance, are often enhanced in haloalkaline habitats 42 such as our natural shoreline i. high salinity and humidity. More relevant to our study was the decreased presence of the contamination-resistant Sphingomonas 43 , and of the potential pathogen Arsenophonus 37 , The bacteria enriched in agricultural and semi-natural landscapes are common in environments associated with plants e. soils and roots. Resistance to environmental contamination has been reported or suspected for some of these bacteria. Gammaproteobacteria, enriched in agricultural colonies, are highly adaptable chemotrophs suggested to resist unfavourable conditions 42 , while the Sphingomonas and Gemmatimonas genera in semi-natural hives enriched and present respectively are key bacteria in cadmium-contaminated and saline-alkaline stressed soils Increased Gemmatimonas abundance has also been linked to Pyraclostrobin fungicide application 44 and to long-term use of organic and inorganic fertilisers Sphingomonas are often associated with plant microbiomes, capable of degrading several recalcitrant substances and common helpers of fungi and plants during metal-degradation of soils 43 , Sphingomonas proliferation in leaf microbiomes has been reported after anti-pathogen treatment of plants A similar Sphingomonas enrichment to the one observed here in semi-natural hives was reported for petroleum-contaminated soils Indeed, semi-natural and agricultural apiaries were situated near traditional oil and natural gas exploitations Enrichment of potential pathogens was another common trait of semi-natural and agricultural hives. Agricultural samples were particularly enriched in Pseudomonas 50 and Arsenophonus. In bees, Arsenophonus has been linked to increased death rates 26 and weakened colony health 37 , whilst enrichment of one Arsenophonus candidate was associated with increased incidence of Colony Collapse Disorder Interestingly, one study showed that both environment and social interactions play an important role in honey bee Arsenophonus acquisition 26 , and Arsenophonus was enriched in all agricultural hive niches. Arsenophonus could be a biomarker of anthropization, transmitted through social activities. However, the same study 26 demonstrated that Arsenophonus abundances within honey bee guts were very location-dependent, with nearby hives sharing similar abundances. Thus, we cannot discard the influence of apiary in the differences detected for Arsenophonus abundances. Semi-natural hives also revealed several potential honey bee pathogens, including the human-affecting Streptococcus 51 , Anaerococcus 52 and Paenibacillus. The latter genus posed a great risk to honey bees, since Paenibacillus larvae is the causative agent of American Foulbrood AFB If transmitted from entrance to bees, Paenibacillus could infect the brood and threaten semi-natural colonies. Likewise, the Lactobacillus kunkeei present in these semi-natural beehives, if transmitted to the brood, could protect semi-natural colonies by increasing brood resistance against both P. larvae and Nosema ceranae 7 , Further studies would be needed to determine P. larvae transmission within beehives. On the contrary, the hive entrances of natural hives exhibited an adapted and overall more pathogen-poor bacterial community, with the exception of P. larvae more abundant than in agriculture. Agricultural hive entrances, compared to semi-natural, had less positive reinforcements against pathogens and contamination i. less abundance of Lactobacillus kunkeei and bioremediators. Whilst natural hive entrances recruited functions prevalent among balanced metabolisms e. methionine synthesis , agricultural hive entrances had more active stress-related pathways. They recruited Gram negative bacterial pathways for synthesis of outer membrane components e. Kdo2-lipid A, LPS as well as the stringent response-inducing ppGpp metabolism, which enables bacterial persistence and pathogenicity 55 , The semi-natural profiles shifted towards natural abundances. Hive internal air and bee bread were the least influenced niches in the apibiome. Internal air, formed by floating abiotic and biotic particles found within hives, was expected to act as an indicator of beehive fitness. Indeed, shifts in airborne microbiome composition have been linked to soil, flora and possibly pollution 57 , as well as urbanisation Consequently, we expected differences between anthropic and natural landscapes, but the internal hive air microbiota turned out to be mostly stable and largely unaffected by environmental factors. This could be because free floating particles within hives, as happens with pollen granules, might stick to bee bodies and reduce the pool of available bacteria that can be sampled from the in-hive air. Anthropization had a meagre effect on bee bread, but differences among environments were consistent with the changes detected in gut and hive entrance. Bee bread sample composition resembled previous studies 8 , 23 , 30 , and overall natural hives had slight enrichment of acidic and sugar-tolerant bacteria while agricultural hives were enriched for Arsenophonus , previously described in bee bread obtained from multiple habitats Herein, Arsenophonus was most likely transmitted from agricultural bees to their food stores or vice versa, as agricultural bee guts possessed slight Arsenophonus enrichment. Contamination of the food reserves by potential pathogens such as Arsenophonus might negatively impact honey bee health at the agricultural environment, as consumption of said food could result in gut microbiome dysbiosis or affect the whole apibiome. One of the main findings of this study was the intermediated relative abundances observed in semi-natural hives for some bacteria e. Comensalibacter , Lactobacillus , Arsenophonus and several predicted pathways e. The intermediate state was more widespread for pathway recruitment than for community composition, indicating an early functional response of the beehive bacterial community. Importantly, 16 days of exposure to an anthropization gradient were sufficient to shift the bacterial fraction of the apibiome of hives. Agricultural hives stayed anthropized while semi-natural bacterial apibiomes became more balanced, resembling the profiles found in a non-anthropized apiary. As a parallel, quick adaptability of the gut microbiome when under pressure has been reported in humans 59 , 60 and other mammals Similarly, honey bee colony-wide analysis of molecular biomarkers also demonstrated an overall increase in the expression of vitellogenin regulatory protein within bees , antioxidant enzymes and immune proteins in hives situated near areas under wildlife recuperation for the US Conservation Reserve Program Our results show that placing beehives in less anthropized areas more natural with less agricultural pressure would lead to recruitment of beneficial bacteria e. Lactobacillus and Commensalibacter in honeybee guts and induce functional reorganisation. Indeed, habitat restoration in agricultural areas by planting native flora can favour recovery of pollinator populations 3 , including wild bees Decreased anthropization of hives increased the relative abundances of beneficial bacteria in all of the sampled hive niches of the apibiome, albeit at different rates, and induced shifts in predicted functional profiles of guts and hive entrances. These results highlight the quick adaptability of honey bee-associated microbiomes. They offer straightforward management strategies to strengthen bee colonies by reducing the impact of anthropization by planting of indigenous flora around crops, or relocating hives to more natural areas whilst maintaining current agricultural production. These results also highlight the relevance of the hive as the unit of study for microbial research, as opposed to bees, in order to understand the contribution of each niche to colony health and resilience, as well as the importance of their interactions. Larger longitudinal and long-term analyses considering seasonal changes would enable the identification of global patterns of anthropization and of core microbes within hive niches, and contribute towards the identification of: 1 beneficial profiles that could be targeted to strengthen honey bee health at any time-period, 2 bacteria that weaken colonies, and 3 biomarkers, as Arsenophonus appears in this work, indicating the risk status of hives under anthropization. Thus, a checklist of safety and hazard markers could be developed as a management tool to employ as a bioindicator of beehive health. Samples were obtained from 3 apiaries within Croatia. On 20 May , 33 hives were formed in the agricultural region of Marijančaci Hives contained one super each. All frames were standard Langstroth, and sister queens and same-origin worker bees were utilised to avoid genetic variation. Hives were moved the next day. Twenty-two hives were relocated 24 aerial km away to the agricultural region of Kozarac The remaining 11 colonies were moved to Vardarac These 11 colonies were designated as the semi-natural apiary. Ten additional hives were located in Unije Seven hives were already established on this natural location beforehand. All 7 had 1 lacked management since including pesticide treatments , 2 survived Varroa destructor infestations and 3 been previously studied by Muñoz-Colmenero et al. Two supers were added to all 10 hives located in this natural landscape. All three apiaries remained untreated during this experiment. Agricultural exploitations and commercial beekeeping practices are regular in Osječko-Baranjska encompassing both Kozarac and Vardarac apiaries. Grasslands, fruit trees apple and plum , and intensive commercial crops such as rapeseed, wheat, sunflower, corn, soybeans and barley surrounded the agricultural apiary 30 , 64 , In contrast, the natural location of Unije had pastureland, tufted hair grass, maquis olive groves , coniferous woodland, and mixed broad-leaved trees holm oaks Arable land was limited to small grassland and shrub plantations around the village Colony strength parameters were assessed on the 6th of June following the Liebefeld method 67 , Portable electronic scales were used to weigh entire hives. Both sides of all frames were checked for in-field measurement of adult bees, brood and pollen areas dm 2 , as well as frame walls and bottom board for measurement of adults. Total adult bee, brood and pollen loads were calculated by multiplying the area by adult bees or brood and pollen as required for standard LR Langstroth frames Varroa load was measured simultaneously by the Powdered Sugar method Statistical analyses for colony strength differences among environments were performed in R Rv3. Samples were collected in June 6, , 17 days after colony formation. When possible, 4 sample types were collected per hive: young worker bees collected from brood frames most likely nurses for gut dissection G , 8 cm 2 of bee bread comb randomly selected from a single frame PB , microorganisms stuck to the hive entrance and collected by swab scrubbing the entire entrance S , and filters containing vacuum filtered internal air F from the hive. Older workers could not be sampled due to the short period elapsed since hive formation. The entrance was swabbed by scrubbing left and right around 6 times per swab tip 3—4 swabs per hive. Sampling of air was done by placing, on top of the honey super, a plastic dome Lekliško cupola, produced by Dubravko Leskovic with a perforated side attached to a vacuum hoover Hf, J. Holdings with filters. The vacuum was left running for 10 min. Air samples were not collected in the semi-natural environment due to material limitations. In total, samples were collected from the 43 hives comprising this study. The supernatant was collected and placed in a clean 1. This process was repeated once more by adding µl 1xPBS to recover the maximum liquid sample size. Cell lysis and DNA extraction was performed following the protocol established by Muñoz-Colmenero et al. Then the tubes were vortexed and incubated at 56 °C and rpm for 90 min. The resulting supernatant was collected and placed in a clean 2 mL tube. This step was performed twice in order to recover as many microorganisms as possible, after which both supernatants were combined and μL of ethanol were added. A 2 mL PowerBead Tube with 0. Samples were homogenised using a Precellys 24 tissue homogenizer Bertin Technologies for 4 min. Then the tubes were incubated at 65 °C for 15 min and centrifuged at 10,× g for 30 s. The supernatant was collected and combined with μL of solution C2, after which tubes were again incubated at 4 °C for 5 min and centrifuged at 10,× g for 1 min. Approximately μL of supernatant were then transferred to a 2 ml collection tube, where μL of pre-shaken solution C4 were added. These primers contained Illumina sequencing adaptors and a 12 bp barcode sequence bound to the forward primers, allowing sample identification. PCR products were examined on a 1. The DNA purification of the PCR products, the preparation of the libraries and the paired-end sequencing were performed at the Sequencing and Genotyping Unit of the University of the Basque Country SGIKER. Quality of raw sequences was checked with FastQC High Throughput Sequence QC Report v0. Demultiplexing of the sequences without Golay error correction , in-depth sequence quality control by the denoise-paired DADA2 method 72 , and taxonomic assignment were performed in Qiime2 v2. Amplicon sequence variants ASVs present in a single sample were removed. The original feature table was split by sample type to create sample type specific datasets gut, bee bread, hive entrance and internal air datasets. Phylogenetic trees were generated from these datasets using mafft and fasttree alignment in Qiime2. Taxonomic analysis was preceded by mitochondrial and chloroplast sequence removal. Relative abundances of bacteria were represented for phyla and genera via qiime taxa barplot. A common sequencing depth for all sample types was determined through alpha rarefaction curves, and utilised to calculate and compare Alpha diversity values among sample types. Samples presenting lower sequence depths were thus filtered out. Visualisation was conducted in R with ggplot2 and dplyr. Sequencing depth for each sample type was then determined through alpha rarefaction curves, and rarefied data sets were obtained per sample type for comparison of environments through Alpha and beta Diversity analyses. For each hive niche, Alpha diversity analyses were performed again and for Beta diversity analyses Bray—Curtis distance community composition dissimilarity was computed using Qiime2 and visualised as Principal Coordinate Analysis PCoA via Vega editor v5. Permutational multivariate analysis of variance PERMANOVA was calculated in Qiime2 based on rarefied Bray—Curtis matrices and with pairwise BH-FDR correction, to determine whether the bacterial communities between environments differed significantly. Considering that data dispersion can confound PERMANOVA results, homogeneity of group dispersion PERMDISP 77 for environments was calculated with betadisper on the same matrices. Spearman correlation-based circular UPGMA trees unweighted pair group method with arithmetic mean were obtained in Qiime2 and displayed via iTOL Interactive Tree of Life tool, v6. Colors indicating anthropization level within UPGMA trees were added via INKSCAPE v0. The feature frequency tables of each sample type were collapsed at genus level and transformed to relative abundances. Tables were uploaded to the Galaxy web application 79 where Linear Discrimination Analysis LDA size Effect LEfSe 80 was used to identify the bacteria driving the differences among environments. LEfSe uses a non-parametric factorial Kruskal—Wallis sum rank test 81 to identify differentially abundant taxa, followed by a canonical method to calculate which taxa combinations contribute more to environmental differences. Histograms and cladograms of results were plotted within the Galaxy web application, and taxa names within graphs cleaned using INKSCAPE v0. test from the Hmisc package, applying BH-FDR correction, and visualized using corrplot package. Non-linear sample distribution was checked before Spearman correlation analysis, using shapiro. test normality test in R 82 and BH-FDR correction. Mean relative abundances of all significant genera were represented in tables using percentages. To determine if environmental changes could impact honey bee apibiome functionality, functional prediction of E. enzymes 83 and MetaCyc pathways 84 were performed for gut and hive entrance samples using the PICRUSt2 v2. The resulting E. and pathway tables were rarefied for diversity analyses. BH-FDR correction was applied to p-values of pairwise analysis. Bray—Curtis distances were visualised via PCoA to determine environmental dissimilarities. Mean relative abundances of significant features were calculated and visualised as histograms via the ggplot2 and dplyr R packages. Kulhanek, K. et al. A national survey of managed honey bee — annual colony losses in the USA. Article Google Scholar. Potts, S. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Li, G. The wisdom of honeybee defenses against environmental stresses. Horak, R. Symbionts shape host innate immunity in honeybees. Dosch, C. The gut microbiota can provide viral tolerance in the honey bee. Article CAS Google Scholar. Emery, O. Immune system stimulation by the gut symbiont Frischella perrara in the honey bee Apis mellifera. Forsgren, E. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 41 1 , 99— Anderson, K. An emerging paradigm of colony health: Microbial balance of the honey bee and hive Apis mellifera. Insectes Soc. Zheng, H. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Article ADS CAS Google Scholar. Kešnerová, L. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. Bonilla-Rosso, G. Functional roles and metabolic niches in the honey bee gut microbiota. Ellegaard, K. Extensive intra-phylotype diversity in lactobacilli and bifidobacteria from the honeybee gut. BMC Genomics 16 1 , Kwong, W. Gut microbial communities of social bees. Jones, J. Gut microbiota composition is associated with environmental landscape in honey bees. The impact of winter feed type on intestinal microbiota and parasites in honey bees. Apidologie 49 2 , — Castelli, L. Impact of nutritional stress on honeybee gut microbiota, immunity, and Nosema ceranae infection. Campbell, J. The fungicide Pristine ® inhibits mitochondrial function in vitro but not flight metabolic rates in honey bees. Kakumanu, M. Honey bee gut microbiome is altered by in-hive pesticide exposures. Abbo, P. Effects of Imidacloprid and Varroa destructor on survival and health of European honey bees, Apis mellifera. Insect Sci. DeGrandi-Hoffman, G. Honey bee gut microbial communities are robust to the fungicide Pristine ® consumed in pollen. Apidologie 48 3 , — Motta, E. Prospects for probiotics in social bees. Corby-Harris, V. The bacterial communities associated with honey bee Apis mellifera foragers. PLoS ONE 9 4 , e Donkersley, P. Bacterial communities associated with honeybee food stores are correlated with land use. Mullin, C. High levels of miticides and agrochemicals in North American apiaries: Implications for honey bee health. PLoS ONE 5 3 , e Miller, D. Transitions and transmission: Behavior and physiology as drivers of honey bee-associated microbial communities. Drew, G. Transitions in symbiosis: Evidence for environmental acquisition and social transmission within a clade of heritable symbionts. ISME J. Forfert, N. Parasites and pathogens of the honeybee Apis mellifera and their influence on inter-colonial transmission. PLoS ONE 10 10 , e Erban, T. Bacterial community associated with worker honeybees Apis mellifera affected by European foulbrood. PeerJ 5 , e Microbial ecology of the hive and pollination landscape: Bacterial associates from floral nectar, the alimentary tract and stored food of honey bees Apis mellifera. PLoS ONE 8 12 , e Muñoz-Colmenero, M. Differences in honey bee bacterial diversity and composition in agricultural and pristine environments—A field study. Apidologie 51 6 , — Locke, B. Characteristics of honey bee colonies Apis mellifera in Sweden surviving Varroa destructor infestation. Apidologie 42 , Gut microbiota structure differs between honeybees in winter and summer. Raymann, K. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. Martinson, V. Establishment of characteristic gut bacteria during development of the honeybee worker. Immune system stimulation by the native gut microbiota of honey bees. Koch, H. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Article ADS Google Scholar. Budge, G. Identifying bacterial predictors of honey bee health. Cornman, R. Pathogen webs in collapsing honey bee colonies. PLoS ONE 7 8 , e Genomics and host specialization of honey bee and bumble bee gut symbionts. Raza, M. Gut microbiota promotes host resistance to low-temperature stress by stimulating its arginine and proline metabolism pathway in adult Bactrocera dorsalis. PLoS Pathog. Kaivosaari, S. N-glucuronidation of drugs and other xenobiotics by human and animal UDP-glucuronosyltransferases. In this case, nutrient loads are low compared with the environments discussed above and the ecosystem is far from being eutrophicated. However, as in the more complex environments Zhang et al. At times when perturbation was higher i. summer , the composition of bacterial communities deviated clearly from that of the reference sites: a high proportion of sequences from the Gammaproteobacteria and Bacteroidetes , as well as a reduction of Alphaproteobacteria , was observed; typical oligotrophic groups such as SAR11 or SAR86 decreased or disappeared, and the main alphaproteobacterial species belonged to the Roseobacter clade Nogales et al. These patterns are the same as those found in VH and the Venice lagoon Zhang et al. Therefore, independently of the particular phylotypes observed in each site which appeared to be different after sequence comparison , there seems to be a general response of microbial communities to nutrient enrichment, at least in seawater, which seems to be independent of the absolute magnitude of the nutrient load. Thus, there seems to be a counter-selection against typical oligotrophic microorganisms and the proliferation of copiotrophic bacteria able to grow and respond to increased nutrient concentrations, such as certain groups of Gammaproteobacteria i. Alteromonadales , Pseudoalteromonas , Pseudomonadales , OM60 group , members of the division Bacteroidetes and representatives of the Roseobacter clade in the Alphaproteobacteria. All these groups have been associated with experimental nutrient enrichment or algal blooms in marine environments Eilers et al. These microorganisms probably react directly to nutrient enrichment by increasing their growth rate and their biomass, following the typical strategy of fast growers. None of the studies conducted so far have explored at the same time the diversity of phytoplankton and bacterioplankton in nutrient-enriched environments, and, therefore, this aspect remains unknown. A good model for studying more directly the effect of nutrient enrichment in marine ecosystems without the presence of additional stressors is aquaculture. Fish production in aquaculture farms is a growing economical sector worldwide but has the drawback of releasing high amounts of organic matter resulting from uneaten fish food and faecal material to the seawater and the sediments below the cages. Because aquaculture has been shown to have an effect on environmental quality and health, there are numerous studies addressing this issue. Meta-analysis of published ecological data from the water column, as well as studies conducted in particular locations, have shown that fish farms have a significant local effect on the pools of particulate and dissolved organic and inorganic nutrients, such as particulate organic phosphorus, particulate organic nitrogen, dissolved organic nitrogen, dissolved organic carbon, ammonium and nitrite La Rosa et al. Usually, no effects are observed on the concentration of phosphate and silicate. As in most complex environments nutrient enrichment from fish cages causes a significant increase in bacterioplankton abundance and heterotrophic production Sakami et al. Although increases in phytoplankton abundance is not always evident from chlorophyll data, a stimulatory effect of fish cages on autotrophic eukaryotic phytoplankton has been observed as well, although this does not lead to the development of phytoplankton blooms because there seem to be a tight control by grazing Navarro et al. Hydrology is probably also playing a role because fish farms are usually located in areas with good water exchange. Despite all these important perturbations of the marine microbial food web, few studies have reported which are the changes in microbial communities that occur under fish cages in comparison with control areas. In a study performed in farms of milkfish Chanos chanos in the Philippines, Garren et al. There were few phylogenetic groups represented in libraries from the fish pens and the proportion of clones affiliated to the Cyanobacteria , the Proteobacteria and the Bacteroidetes were high. The results of this study would indicate a reduction in the bacterioplankton diversity near fish cages in comparison with surrounding areas, contradicting the findings reported above about microbial diversity in nutrient-enriched coastal areas. This apparent reduction in bacterial diversity due to aquaculture should be confirmed in other systems because it might not be a general finding. For example, Wei et al. However, it should be taken into account that fish pens and ponds are completely different aquaculture systems and therefore the effect on microbial communities might also be different. On the other hand, if we take into account the fact that the perturbation caused by a fish farm is more constant in amount, composition and periodicity of additions i. fish food and fish faeces , we can hypothesize that this might lead to the development of microbial communities highly specialized in processing this particular type of organic load, maybe dominated by a few very efficient microorganisms, and therefore less diverse. The most dramatic effect of the nutrient load from fish farms occurs in sediments below the cages. There, the high organic load, mainly in the form of particulate organic matter, increases oxygen demand and alters the biogeochemistry of the sediments. The benthic biomass in environments affected by fish farms becomes dominated by microbial components, and bacterial abundance increases significantly compared with control areas Mirto et al. In contrast, fish farms cause a decline of some groups of benthic fauna and seagrasses Mirto et al. With respect to functionality, the organic load of fish farming stimulates microbial anaerobic respiration processes in sediments below fish farms, such as sulphate, iron and manganese reduction, denitrification, methanogenesis and fermentation Christensen et al. By far the most important anaerobic process in fish-farm sediments, as in other nutrient-enriched environments, is sulphate reduction because sulphate is usually present in nonlimiting concentrations in marine sediments Holmer et al. The production of high amounts of hydrogen sulphide by sulphate reduction, concomitant with a decrease in oxygen, might lead to the inhibition of important microbial processes in the nitrogen cycle such as nitrification oxidation of ammonium to nitrate , and hence, to lower the rates of coupled denitrification reduction of nitrate to nitrogen gas , which requires oxidized forms of nitrogen. Coupled nitrification—denitrification is important for reducing the high nitrogen load of fish-farm sediments and for controlling the efflux of ammonium from the sediments to the water column McCaig et al. However, in reduced fish-farm sediments, the process of dissimilatory nitrate reduction to ammonium, which also needs oxidized forms of nitrogen, can be quantitatively more important than denitrification. As a result of reduced nitrification, there is less nitrogen removed from the sediment as nitrogen gas ultimately released to the atmosphere and a higher efflux of ammonium to the water column, which stimulates primary production and maintains the nitrogen in the system Christensen et al. But even if nitrification is not inhibited, it might be insufficient to oxidize all the ammonium resulting from the organic load to nitrate, and consequently there is a net flux of ammonium to the water column Bissett et al. The bacterial composition in sediments under fish farms along gradients of organic pollution, and hence sulphide and oxygen concentrations, have been analysed by 16S rRNA gene-based approaches in Japan Asami et al. Significant differences in composition of clone libraries from fish-farm and reference sediments are usually observed, both at a broad and at a fine phylogenetic resolution Bissett et al. These studies have also shown that bacterial community composition changes significantly after cessation of production, such as in periods of fallowing, but the communities do not revert to the composition before the impact, i. they are not resilient Bissett et al. A distinctive characteristic composition of bacterial communities in fish-farm sediments cannot be easily drawn due to the high microbial diversity found in marine sediments Asami et al. However, even when these farm sediments were located in different areas and they were dedicated to the production of different fish species, there were some common findings that allow the formulation of certain general trends. For example, an increase in the abundance of Deltaproteobacteria which includes most of the sulphate-reducing genera is generally observed. This has been proven by a higher proportion of deltaproteobacterial 16S rRNA gene clones in fish-farm libraries Castine et al. Besides, in agreement with a high production of sulphide by sulphate reduction, a characteristically high proportion of clone sequences of putative sulphur-oxidizing Gammaproteobacteria Asami et al. Sequences of the Epsilonproteobacteria are also abundant Bissett et al. This group is also related to the metabolism of reduced sulphur compounds and it is usually found in sulphide-rich sediments Campbell et al. The proportion of clones affiliated to the division Bacteroidetes Flavobacteria in particular , which are supposed to play a role in biopolymer degradation, tends to be high in fish-farm sediments as well, and the phylotypes recovered are different from those in control areas Asami et al. Finally, clone sequences related to betaproteobacterial nitrifiers are usually not recovered in libraries from fish-farm sediments targeting total bacterial diversity Bissett et al. However, using molecular tools targeting selectively this bacterial group, several authors have demonstrated a different composition of the betaproteobacterial nitrifiers in fish-farm sediments compared with control sites McCaig et al. These changes in the composition of microorganisms participating in the degradation of the organic load and in the sulphur and nitrogen cycles, together with the changes in process rates i. sulphate reduction , provide good evidence of an altered functionality of the microbial communities in sediments below fish farms. The perturbation is likely to have an effect on archaeal groups too, but the composition of archaeal communities or their role in biogeochemical processes such as ammonia oxidation or methanogenesis in these environments has not been explored so far. Taking together the results of the studies conducted in environments receiving point and diffuse nutrient inputs, some general effects of nutrient enrichment on microbial community composition and function can be proposed Fig. Thus, despite the composition of the microbial assemblage developing at a particular site, it seems clear that the structure of the microbial food web will be affected as well as important biogeochemical cycles, such as the nitrogen, carbon and sulphur cycles. General effects of two examples of human-derived stressors on marine microbial communities. Persistent organic pollutants, such as herbicides aldrin, DDT, hexachlorobenzene, etc. They reach the marine environment by direct discharges from coastal areas or dumping into the sea , runoff from land, river discharges or atmospheric deposition. They also adsorb to floating and stranded plastics, which contributes to their transport and persistence in the environment Rios et al. Among the different pollutants entering the marine environment, the effect of hydrocarbon pollution on marine microbial communities has been the subject of numerous studies, mainly driven by the concerns caused by tanker accidents such as that of the Exxon Valdez in Alaska or the more recent accident of the Prestige tanker in Spain. Most of these studies have focused on analysing the effect of crude oil spills on microbial diversity, the changes in response to bioremediation trials usually involving the addition of nutrients and the extent of hydrocarbon biodegradation in polluted environments. These topics are covered in excellent recent review articles Head et al. The results obtained show that the microbial communities developing in response to events of hydrocarbon contamination differ in composition although the efficacy in hydrocarbon removal is similar Head et al. Events of acute contamination cause a reduction of microbial diversity in the short term. This is due to two main reasons: the disappearance of certain groups of microorganisms i. archaea and cyanobacteria and the strong selection for specialist hydrocarbon-degrading marine bacteria i. Alcanivorax , Cycloclasticus , which become predominant particularly when nutrients are added to stimulate hydrocarbon degradation Head et al. In contrast, microbial diversity is usually high in chronically or long-term hydrocarbon-polluted marine environments Hernandez-Raquet et al. This behaviour is exactly the same as that observed in environments polluted chronically with nutrients Zhang et al. Typically, 16S rRNA gene sequences related to known hydrocarbon degraders [i. Alcanivorax , Cycloclasticus , etc. Yakimov et al. This might indicate that those hydrocarbon degraders are minor components of these communities, and therefore are missed because of method limitation, although they become predominant in bioremediation trials. For example, sequences affiliated to Alcanivorax were not observed in mesocosms prepared with seawater from Messina Harbour but they became predominant 15 days after the addition of oil and nutrients Cappello et al. Also, most-probable-number counts of hydrocarbon degraders in two marinas in the United States were shown to be low Piehler et al. Recent studies conducted in areas polluted after the Prestige tanker oil spill sampled one year after the accident revealed the importance of members of the suborder Corynebacterineae i. Rhodococcus and the family Sphingomonadaceae in the degradation of the alkane and aromatic fraction of this heavy oil, respectively Alonso-Gutiérrez et al. These results show the wide diversity of bacterial hydrocarbon degraders in environmental samples and the importance of certain groups i. Actinobacteria as hydrocarbon degraders in rough environments i. rocks polluted with heavy oil. Alternatively, there might be novel, uncharacterized degraders in these polluted environments Paissé et al. We have experimental evidence supporting this hypothesis coming from an analysis of aromatic ring-hydroxylating dioxygenase genes ARHD in polluted coastal sediments from Patagonia in Argentina Lozada et al. Sequences representative of the already known phnAc -like genes of Alcaligenes faecalis AFK2, phnAI -like genes of Cycloclasticus spp. and nahAc -like genes of Pseudomonas spp. These novel ARHDs contained the conserved residues of bacterial ring-hydroxylating dioxygenase α-subunits Lozada et al. We have a biased perception of the risk associated to hydrocarbon pollution towards contamination caused by tanker accidents and this has motivated most of the research on hydrocarbon pollution and bioremediation studies conducted so far. However, the contribution of tanker accidents to total hydrocarbon contamination in the sea is very low. For example, in the Mediterranean Sea, 80 tonnes of oil were spilled in accidents between and [ Environmental European Agency EEA , ]. In contrast, tonnes are discharged per year due to shipping operations i. discharges of ballast water, tank washing, oil sludge, bilge water or engine room wastes and about tonnes per year from oil terminals and routine land-based operations EEA, Therefore, it is important to take into account this type of pollution and to assess properly its effects on coastal microbial communities. An example of a study of the effect of land-based sources of hydrocarbon pollution on microbial communities is the one done in a chronically polluted coastal retention basin in the Mediterranean Sea Etang de Berre, France. This coastal lagoon receives hydrocarbons from refineries, petrochemical plants and transportation systems. Samples were taken along the hydrocarbon pollution gradient Hernandez-Raquet et al. This means that, although other environmental factors were determining microbial composition in the area, hydrocarbons appeared as a key factor for diversification of communities. When the composition of bacterial communities was analysed in detail, a predominance of sequences related to Delta - and Gammaproteobacteria , as usually observed in marine sediments, was obtained, although variability in different sampling years was high as happens also in nutrient-enriched environments Hernandez-Raquet et al. As usual, only a few sequences could be related to hydrocarbon-degrading bacteria and these included sequences related to Marinobacter spp. Therefore, anaerobic metabolism of hydrocarbons by SRB seems to be common in polluted environments. As mentioned above, activities derived from shipping are important sources of hydrocarbon contamination in the marine environment EEA, ; Halpern et al. A good proportion of the pollution caused by ships is due to illegal discharges. In the Mediterranean, for example, the density of spills can be correlated with major shipping routes Ferraro et al. The effect of this type of diffuse hydrocarbon pollution in the composition of seawater microbial communities has not been addressed but it might be a factor to consider for explaining the described cosmopolitan distribution of specialist hydrocarbon degraders in the marine environment Yakimov et al. In areas exploited for tourism, there is a consistent increase in the number of hydrocarbon spill detections during the high season, correlating with the increase in boating activity Ferraro et al. The pollution caused by maritime transport and recreational boats is likely to be a source of fuels i. diesel and synthetic lubricants to seawater and sediments. There are few studies addressing the effect of this type of pollution. One experiment was performed in in situ mesocosms with water and sediments from a tropical estuary in Singapore, impacted by boating-derived activities. The mesocosms were treated with diesel oil at concentrations reproducing the mean and the highest hydrocarbon concentrations measured in the area Nayar et al. The response observed was negative for eukaryotic phytoplankton and picocyanobacteria Synechococcus at higher hydrocarbon concentrations, although picocyanobacteria were stimulated at the lower concentrations. In contrast, the number of heterotrophic bacteria and production rates increased in response to the treatment, particularly at the higher concentrations Nayar et al. These results show that there is a rapid response of microbial communities to diesel pollution in the marine environment, as demonstrated for more complex hydrocarbon mixtures such as crude oil Head et al. A second study tested the short-term effect of diesel oil, a biodegradable lubricant and a synthetic lubricant clean and used in a field experiment in pristine Antarctic sediments Powell et al. Although the bacterial communities differed from the control in all treatments, the most significant variations were due to treatment with diesel, followed by synthetic lubricants, but not with the biodegradable lubricant Powell et al. These results show ways of reducing the impact of lubricants in the marine environment, particularly in sensitive areas such as the poles. Maritime activities require the building of ports and recreational marinas. Because of the constant increase of maritime transport United Nations Conference on Trade and Development, and the interest in increasing tourism-derived activities European Commission, , there is a constant pressure for the development of harbours and recreational marinas, and therefore, we can expect an increase in the environmental stress posed by them. Some of the pollutants that are commonly found in harbours are hydrocarbons i. as result of boat traffic, accidental spills or discharge of bilge oil and ballast water , detergents, surfactants or antifouling compounds i. heavy metals, biocides , even though ports have facilities for the collection of wastes and for pollution control. Additionally, they receive nutrient inputs, either directly due to harbour activities or through discharges of rivers and sewage effluents such as VH in Hong Kong Zhang et al. Harbour seawater and sediments have been shown to hold particular bacterial communities, different from those in adjacent areas, highly diverse and highly variable at the temporal scale Schauer et al. Despite chemical pollution, the effect of nutrient enrichment seems to be the most important factor driving the composition of bacterioplankton in harbours, and for example nutrient enrichment might explain the high abundance of sequences related to Clostridium and Vibrio in the sediments of Milazzo Harbour, subjected to high organic load Yakimov et al. Relationships between bacterioplankton community composition and hydrocarbon pollution were only found in the study conducted in Messina Harbour, where the relative abundance of putative hydrocarbon-degrading bacteria correlated with an increase in hydrocarbons Denaro et al. In sediments, stimulation of hydrocarbon degraders has been observed after hydrocarbon addition, indicating that there is a primed microbial community able to use these pollutants, both aerobically and anaerobically Hayes et al. As in other hydrocarbon-polluted sediments, SRB seem to be involved in hydrocarbon degradation in harbour sediments. For example, a recent study on the composition of the active sulphate-reducing assemblages in sediments of Boston Harbour by analysing transcripts mRNAs of dsrAB genes Chin et al. Several studies have addressed the composition of bacterial communities in marine sediments polluted with heavy metals. As in the case of other pollutants, bacterial communities in sediments with different levels of heavy-metal pollution are shown to differ Gillan et al. The same result has been observed for archaeal and photosynthetic communities Toes et al. Different from the cases of nutrient or hydrocarbon pollution, which seem to stimulate bacterial growth, total prokaryotic cell numbers appear to be negatively correlated with heavy metals such as cadmium, copper, zinc and lead Gillan et al. However, when the calculations were done for particular phylogenetic groups, few statistically significant correlations could be established, and they seemed to be dependent on the sample. Therefore, it is difficult to determine the effect of heavy metal on particular groups of microorganisms with the data available so far. In highly polluted sediments of a Norwegian fjord, the abundance of Gammaproteobacteria determined by FISH counts was negatively correlated with copper and zinc, and that of Bacteroidetes with copper, zinc and cadmium. In contrast, in Belgian sediments with lower pollution levels, the only significant correlation was found between Bacteroidetes and cadmium Gillan et al. In these two sediments, bacterial community composition was also analysed by a cloning and sequencing approach. The results evidenced the presence of the main groups typically found in marine sediments: Gamma - and Deltaproteobacteria and Bacteroidetes. But some of the clones in the Norwegian sediment grouped with clones from an Antarctic sediment polluted with heavy metals and hydrocarbons, and a group of gammaproteobacterial sequences was found in both the Norwegian and the Belgian sediments Gillan et al. This might indicate that certain bacterial populations might be favoured in heavy-metal-polluted sediments, although these relationships are even more difficult to draw than in the case of hydrocarbon pollution. A laboratory experiment was performed in microcosms to simulate the effect of sediment disturbance dredging in a sediment chronically polluted with heavy metals. In addition, the effect of deposition of a 3 mm layer of polluted sediment on the surface of unpolluted sandy sediments was also analysed Toes et al. These two practices dredging and deposition onto a different location are common in the marine environment. Homogenization of the polluted sediment simulating dredging caused minor changes in the composition of microbial communities. However, overlying of sandy sediments with polluted sediment caused significant changes in the composition in the three microbial components analysed Bacteria , Archaea and Cyanobacteria in comparison with the unpolluted control, although the effect was lower for Archaea Toes et al. After one year of incubation, some common bacterial groups were observed in clone libraries from the artificially polluted sandy sediments in comparison with those from the chronically polluted sediment: representatives of the Roseobacter clade, the genus Vibrio and a member of the Flavobacteriaceae whose sequence was related to a clone from a heavy-metal-polluted Antarctic sediment Toes et al. These sequences might represent microorganisms favoured by the conditions imposed by heavy-metal pollution, although to confirm this, it will be necessary to corroborate their absence or lower abundance in control, unpolluted sediments. In the environment, removal of sediments polluted with mercury, PCBs and PAHs in a location in the Baltic Sea resulted in the development of significantly different bacterial communities. Summarizing the studies presented on chemical and heavy-metal pollution, we can conclude that, as in the case of nutrient enrichment, this type of perturbation again alters the composition of microbial communities Fig. But because this type of pollution selects for those microorganisms that are capable of degrading or chemically transforming the pollutants, and sometimes these organisms belong to particular phylogenetic groups, the studies conducted so far can point to particular types of bacteria i. oligotrophic marine hydrocarbon-degrading Gammaproteobacteria , Actinobacteria , Sphingomonaceae or certain SRB phylotypes likely to be contributing to alleviate the problem of chemical pollution in the environment. However, in many cases, these is only indirect evidence that does not fill the gap in knowledge on fundamental aspects of the functionality of microbial communities in chemically polluted environments. For example, novel or unrecognized microbial genera might be involved in the degradation of contaminants, most likely through metabolic cooperation with other microorganisms using degradation intermediates. Microorganisms as such, i. forming part of degradation networks, will not be isolated as typical pollutant degraders and therefore escape our knowledge, although they are probably key players in the environment. Other fundamental aspects, such as how pollution affects bacterial production rates, respiration rates or other microbial processes have not been analysed in chemically polluted environments, in contrast to what has been done in nutrient-enriched environments. The broad use of high amounts of antibiotics in intensive farming to prevent or treat animal infections is also causing environmental concern see review Cabello et al. The bactericidal action of antibiotics can cause changes in the composition of natural microbial communities by selectively inhibiting susceptible bacteria. Besides, exposure of natural communities to antibiotics might lead to the selection of resistant bacteria, which can then transfer their resistance determinants to opportunistic pathogenic bacteria in the environment. In relation to the marine environment, there are numerous data coming from aquaculture, where administration of high amounts of a variety of antibiotics in fish farms for prophylactic or therapeutic reasons is routinely done Cabello, ; Sapkota et al. Antibiotic treatment in fish farms leads to increased concentrations of these compounds in water and sediments below the cages as well as in the fish stock produced. As a result, there is a positive selection for antibiotic-resistant bacteria in water and sediments from fish cages, surrounding areas and among fish-associated bacteria Chelossi et al. Often, the isolation of multiresistant bacteria i. bacteria resistant to several antibiotics is reported. This is in agreement with the simultaneous use of several types of antibiotics in fish farms Dang et al. Antibiotic-resistant bacteria isolated from fish-farm water and sediments are diverse. Although most of them belong to the Gammaproteobacteria genera Vibrio , Photobacterium , Pseudomonas , Pseudoalteromonas , Alteromonas , Citrobacter , Salmonella , isolates from the Alphaproteobacteria , Firmicutes , Actinobacteria and Bacteroidetes have also been reported Furushita et al. The presence of antibiotic-resistance genes in these isolates, as well as the ability of some of the isolates for transferring the resistance genes by conjugation to Escherichia coli recipient strains, have been demonstrated Rhodes et al. Besides, closely related plasmids of the IncU group, carrying oxytetracycline-resistance determinants, have been found in isolates from hospitals in the UK and Germany and from fish-farm environments, demonstrating that plasmid transfer might occur between natural bacteria and potential human pathogens Rhodes et al. Instead of isolating antibiotic-resistant bacteria, Hargrave et al. They measured profiles of resistance to the antibiotic oxytetracycline, widely used in aquaculture, in mixed bacterial communities from sediments around salmon aquaculture farms and in feed pellets which were proposed as the source of antibiotics. Usually, numbers were higher in surface sediments but resistance levels were still high at intermediate depths, which mean that sediments around fish farms can constitute reservoirs for antibiotic-resistant bacteria in the marine environment Hargrave et al. It is important to recognize that fish farms are not the only source of antibiotics in the marine environment. Thus, oxytetracycline-resistant bacteria are present in surface seawater and sediments receiving the input of sewage treatment plants, as demonstrated in Jiaozhou Bay in China and Halifax Harbour in Canada Dang et al. The study conducted in the area of VH, where discharges of Anthropogenic use of coastal areas i. beaches also seems to have an effect on the natural abundance of antibiotic-resistant bacteria. Studies performed in sand from beaches of the Southern Baltic Sea coast showed that bacteria isolated from a recreational beach had higher resistance to antibiotics from different chemical families than those in nonrecreational areas Mudryk, ; Mudryk et al. The level of multiresistance resistance to several antibiotics was also higher among isolates from the recreational beach. The examples presented here provide evidence of the potential health risk derived from antibiotic contamination in the marine environment because they demonstrate the selection and enrichment of multiresistant bacteria. Some of them might be potential human pathogens i. Vibrio , Pseudomonas , Salmonella or might potentially transfer genetic determinants of resistance to pathogenic bacteria. But none of these studies have addressed how antibiotics alter the composition and functionality of microbial communities in marine environments. Given the magnitude and worldwide extension of the problem of antibiotic pollution, it is imperative to design carefully environmental and laboratory studies to address this issue, to determine which microbial groups are affected by the presence of antibiotics in the water and how antibiotics alter the functionality of microbial communities, both in water and in sediments. Several of the uses of coastal resources have the risk of introducing potential human pathogenic bacteria, as shown in Table 1. The highest risk is derived from direct sewage discharges, which are a source of human faecal bacteria. Therefore, sewage treatment and routine controls for faecal indicators coliforms, E. coli , enterococci in seawater in areas dedicated to bathing are usual procedures in developed countries Stewart et al. However, sewage discharge is not the only source of faecal bacteria in marine environments. Other human-derived sources of potential pathogens are runoff from land urban and agricultural areas , leaking septic tanks, sewer overflows, discharges from boats, etc. Stewart et al. Sources of faecal bacteria other than human i. birds, pets, wildlife, etc. should not be disregarded Choi et al. A recent publication has presented a novel and interesting approach to assess faecal pollution in coastal watersheds using community-based indicators Wu et al. Using PhyloChip, a phylogenetic microarray, these authors identified operational taxonomic units OTUs characteristic of faecal samples mainly belonging to the phyla Firmicutes , Proteobacteria , Bacteroidetes and Actinobacteria which were designated as faecal subsample associated OTUs FSAO. By analysing the similarity of bacterial communities in several environmental samples from two watersheds in Santa Barbara CA to FSAO, they could identify the samples exposed to faecal sources. The results obtained agree with traditional culture methods for the detection of faecal bacteria. In addition, they proposed a new community-based indicator to assess ecosystem health, based in the ratio of relative richness of three bacterial classes: Bacilli , Bacteroidetes and Clostridia to that of the Alphaproteobacteria BBC: A. This ratio is higher in faecal and sewage samples and lower in samples not impacted by faecal material Wu et al. Approaches like this represent an important step forward in the analysis of faecal contamination in coastal areas because it is based in powerful molecular methods, and in the assessment of whole communities instead of using a few two or three indicator bacteria. Faecal material is also a source of human pathogenic enteric viruses. Viruses from faecal origin belong to the families Adenoviridae adenovirus , Caliciviridae i. Norwalk virus , Picornaviridae i. poliovirus, hepatitis A and Reoviridae reoviruses and rotaviruses. These viruses cause a variety of diseases in humans, either by direct exposure to water or after ingestion of contaminated seafood Griffin et al. Viruses are shown to persist better in the environment than the bacterial indicators used for water quality monitoring Griffin et al. Apart from causing diseases in human and other marine mammals, epidemic viral infections can cause significant economic losses in aquaculture plants for commercial production of fish and shellfish Lang et al. In addition to discharges of faecal material, other human activities such as bathing constitute a source of potential microbial contamination. For example, enterococci and Staphylococcus aureus are transferred directly from the skin of bathers in high loads into seawater 3—6 × 10 5 and 6. These results highlight the importance of nonenteric bacteria as potential pathogens in the environment Stewart et al. Besides the health risk posed by allochthonous microorganisms from faecal or other origin, there are several potential human pathogenic species that are indigenous to marine and estuarine environments, such as Vibrio cholerae , Vibrio vulnificus and Vibrio parahaemolyticus Thompson et al. The first two species, V. chlolerae and V. vulnificus , can cause severe, fatal diseases. In contrast, V. parahaemolyticus infections are usually not life-threatening but they are very common worldwide, mainly due to the consumption of contaminated seafood Collins et al. These opportunistic pathogenic bacteria have a variety of virulence factors such as haemolysins V. vulnificus and V. parahaemolyticus , toxins cholera toxin , colonization factors, etc. Thompson et al. The presence of pathogenicity-associated genes from V. cholerae and V. parahameolyticus has been proven in environmental isolates from different Vibrio spp. In the marine environment, vibrios are usually associated to surfaces and establish symbiotic relationships with phyto- and zooplankton i. copepods , which might constitute vectors for disease transmission and for the development of disease outbreaks Lipp et al. Therefore, conditions favouring the development of phytoplankton blooms, such as in eutrophicated environments, might favour the development of vibrios. In fact, as mentioned previously in this review, Vibrionaceae have been found in nutrient-rich environments Yakimov et al. In addition, vibrios proliferate at higher water temperatures and have a wide tolerance range to salinity, being able to survive well in low-salinity brackish water. Therefore, their incidence is expected to increase in a scenario of global climate change. Different marine dinoflagellates Dinophysis spp. brevis and Gambierdiscus toxicus and a diatom Pseudonitzschia produce toxins with a variety of neurologic, gastrointestinal, respiratory and irritating effects. Disease is usually caused by consumption of contaminated fish or shellfish, although aerosol inhalation and direct eye or skin exposure can also cause problems. Environmental perturbations derived from human activities that might have an effect in the increased incidence of harmful algal blooms are multiple and include: nutrient enrichment eutrophication , particularly in coastal waters; destruction of coastline due to coastal exploitation i. Potential pathogens might be disseminated in the marine environment due to long-distance transport and discharge of ship ballast water used for ship stability and trim. For example, ballast water has been shown to contain epidemic-causing serotypes of V. coli including strain O , Enterococcus spp. Concerns of the danger of ballast water discharges, not only for the spread of microbial pathogens but also of invasive species, have resulted in the establishment of guidelines for ballast water management and the promulgation of the International Convention for the Control and Management of Ship's Ballast Water and Sediments in by the International Maritime Organization. These numbers are highly variable and depend on many factors such as the source region of the boat, season of the year and ballast-water management, i. whether or not there had been open-ocean exchange of water Drake et al. The changes in the microbial component of ballast water during a transoceanic voyage in tanks with and without open-ocean exchange have been studied by quantifying bacterial and viral numbers Drake et al. The results showed that containment of ballast water in boat tanks led to a decrease in the bacterial and viral load in comparison with the initial water, irrespective of performing open-ocean exchange or not Drake et al. Bacterial communities in ballast water initially resemble those of the source seawater, which is usually from a coastal location Tomaru et al. This ballast water is replaced in the open ocean at some point during the voyage to reduce the risk of discharging invasive species in the reception port. The open-ocean water filling the tanks has a significantly different microbial composition Tomaru et al. This means that allochthonous microorganisms from coastal regions are routinely discharged into the open ocean, and vice versa, open-ocean microorganisms are discharged into coastal regions. However, the microbial assemblages discharged in each case are different from those in the source water because there is a change in composition during the voyage within the time frame of days Tomaru et al. A recent study analysed bacterial diversity in ballast water in a ship anchored in Xiamen Port, and compared it with that of the receiving harbour seawater Ma et al. The source of the ballast water was Singapore and it had been partially replaced with water from the South China Sea. The analysis of the corresponding 16S rRNA gene clone libraries revealed that ballast water contained a less diverse bacterial community, with representatives of only two classes, Alpha - and Gammaproteobacteria. There was no evidence of the presence of pathogenic bacteria. Within these two groups, the phylotypes retrieved in ballast water were different from those in Xiamen Harbour seawater Ma et al. Thus, wherever it is done, the discharge of ballast water might potentially alter, at least transiently, the autochthonous microbial composition of the discharge site. Whether this is significant or not has not been determined. If an open-ocean exchange is performed, we can expect that most of the coastal microorganisms will not be able to compete with the autochthonous bacteria in the more oligotrophic open ocean, and the opposite, i. oligotrophic bacteria from the open ocean will not survive in coastal regions. Results from metagenomic studies show that microbial communities with dissimilar genomic composition i. genetic repertoire develop in different marine locations or at different depths DeLong et al. Therefore, the success of an invasive microorganism, potentially dangerous such as a pathogen, would depend on the size of the inoculum, its capability to survive in the new environment and its competitive ability in facing a microbial community adapted to the conditions at the site of discharge. As reported by Halpern et al. Thus, the interest in studying the effect of processes such as increase of seawater temperature and ocean acidification is increasing rapidly in the last years. No particular studies have been done targeting the microbial compartment but the information gathered so far provides clues on how microbial communities could be affected. Anthropogenic activities such as the burning of fossil fuels, deforestation, industrialization and cement production are causing increased levels of atmospheric carbon dioxide CO 2 and, consequently, an increase in dissolved CO 2 in the oceans. The acidification of the oceans will be detrimental for calcifying organisms and will cause changes in species distribution and abundance, as well as in food-web dynamics and structure. The increase in dissolved CO 2 has an effect in the carbon cycle of the oceans but also in the cycles of the major nutrient elements: nitrogen, phosphorus, silicon and iron for a review, see Hutchings et al. The information available indicates that three important processes in the nitrogen cycle might be affected. For example, nitrogen fixation is predicted to increase in a high-CO 2 ocean, based on the results of experiments done with nitrogen-fixing Trichodesmium and unicellular Cyanobacteria. In contrast, a decrease in pH seems to reduce nitrification rates and the abundance of bacterial and archaeal nitrifiers in seawater Hutchings et al. Consequently, the fluxes of oxidized nitrogen species, such as nitrate, will be affected. This in turn would have a negative effect on denitrification rates, although there can also be positive effects over this process due to the expansion of suboxic zones in the oceans. An increase in the relative proportions of ammonium to nitrate due to decreased nitrification will cause a shift in the composition of primary producers, by favouring microbial components such as picocyanobacteria and nanoflagellates Hutchings et al. On the other hand, global warming will lead to an increase in seawater temperatures and cause a stronger stratification of the oceans. This will limit the fluxes of nutrients from sediments and the deep sea i. phosphorus, iron , and potentially intensify problems of hypoxia and anoxia. Changes in the climate system can also cause alterations in the abundance and global distribution i. spread from tropical areas to lower latitudes of pathogens such as Vibrio spp. The abundance of V. cholerae , the causative agent of cholera epidemics, is already shown to follow climatic patterns Lipp et al. Higher water temperatures, together with increased nutrient levels, would also increase the frequency and severity of harmful algal blooms Moore et al. Environmental changes due to anthropogenic activities seawater warming or increased CO 2 and nutrient supply are also suspected of being involved in the expansion of coral diseases worldwide, diseases in which microorganisms are important Sokolov et al. In addition, strong rainfall events and flooding due to sea-level changes, both expected under a global change scenario, would result in an increase of estuarine and brackish low-salinity environments, which again would favour the growth of Vibrio spp. Lipp et al. Moreover, floods may also cause problems of inefficient water sanitation and increase runoff from land, with the consequent increase in the risk of faecal bacterial and viral , nutrient and chemical contamination of the marine environment Kite-Powell et al. The possibility of generating phytoplankton blooms by ocean fertilization with the aim of increasing the flux of organic matter towards the deep ocean causing carbon burial and alleviating problems of increasing CO 2 concentrations is catching the attention of companies for commercial exploitation. This represents a new anthropogenic use of the marine environment. Several scientific experiments involving iron and phosphorous fertilization have been performed in order to test the premise that these nutrients were limiting primary production in different oceanic regions Thingstad et al. However, based on the experience gathered on the functioning of marine ecosystems and the interplay of biogeochemical cycles, there is strong scientific criticism of the usefulness of ocean fertilization to sequester carbon, as well as concerns about the risks associated Secretariat of the Convention on Biological Diversity, In particular, the plans of the Australian company Ocean Nourishing Corporation to discharge urea in nitrogen-deficient areas of the ocean have been criticized on scientific grounds Glibert et al. In this scenario, there will not be an increase in CO 2 sequestration and carbon burial, and in addition, undesired gases will be released into the atmosphere, aggravating the problem instead of alleviating it. Worldwide marine ecosystems suffer from the impact caused by human activities. In view of the development of our societies and the increase in human population, the stress posed by humans to the marine environment will continue to increase. The microbial component of marine ecosystems has been neglected in many studies of anthropogenic impact. Therefore, our knowledge on microbial communities of anthropogenically impacted environments lags far behind those involving higher organisms. Many studies report changes in prokaryotic numbers and a few have included measurements of microbial activity. Changes in diversity have also been analysed, although in many cases, this has been done only by electrophoretic profiling methods. As a result, we have a partial view on the composition of microbial communities in stressed environments. Therefore, there is a need for a better cataloguing of microbial diversity in anthropogenically stressed environments. New technologies such as remote ocean-observing systems, metagenomics, metatranscriptomics and massive tag sequencing can help us to gain more information on these communities. Thanks to the use of comparative studies of impacted and reference sites, we know that human impact causes significant changes in microbial community composition. However, we are still unable to make predictions on how important this could be for the functioning of the ecosystems, particularly in the long term, especially if the stressor does not disappear or it is likely to increase. For example, we know already that certain microbial groups are favoured in conditions of nutrient enrichment or chemical pollution, but most of this information is provided for broad phylogenetic groups i. division, class and not for particular species or ecotypes. phylum, class, genera, species, etc. There is also a need for relating the changes in diversity to the metabolic processes, which are important for the functioning of the ecosystem. Because of the complexity of marine ecosystems, these goals would need the combination of environmental observations as well as carefully designed microcosms or mesocosms experiments. In the context of global climate change, for which human activities can be blamed, microbial communities of human-stressed environments are of paramount importance, and therefore the study of their composition, dynamics and functioning is of the utmost relevance for the development of less destructive human practices or for alleviating problems already present in our environment. The authors wish to thank Margarita Gomila for her comments on the manuscript. and J. are supported by PhD fellowships of MICINN. Ager D Evans S Li H Lilley AK van der Gast CJ Anthropogenic disturbance affects the structure of bacterial communities. Environ Microbiol 12 : — Google Scholar. Aguiló-Ferretjans MM Bosch R Martín-Cardona C Lalucat J Nogales B Phylogenetic analysis of the composition of bacterial communities in human-exploited coastal environments from Mallorca Island Spain. Syst Appl Microbiol 31 : — Aguirre-Macedo ML Vidal-Martinez VM Herrera-Silveira JA Valdés-Lozano DS Herrera-Rodríguez M Olvera-Novoa MA Ballast water as a vector of coral pathogens in the Gulf of Mexico: the case of the Cayo Arcas coral reef. Mar Pollut Bull 56 : — Allison SD Martiny JBH Resistance, resilience, and redundancy in microbial communities. P Natl Acad Sci USA : — Alonso-Gutiérrez J Figueras A Albaigés J Jiménez N Viñas M Solanas AM Novoa B Bacterial communities from shoreline environments Costa da Morte, Northwestern Spain affected by the Prestige oil spill. Appl Environ Microb 75 : — Alonso-Sáez L Balagué V Sà EL Sánchez O González JM Pinhassi J Massana R Pernthaler J Pedrós-Alió C Gasol JM Seasonality in bacterial diversity in north-west Mediterranean coastal waters: assessment through clone libraries, fingerprinting and FISH. FEMS Microbiol Ecol 60 : 98 — Asami H Aida M Watanabe K Accelerated sulfur cycle in coastal marine sediment beneath areas of intensive shellfish aquaculture. Appl Environ Microb 71 : — Baffone W Tarsi R Pane L Campana R Repetto B Mariottini GL Pruzzo C Detection of free-living and plankton-bound vibrios in coastal waters of the Adriatic Sea Italy and study of their pathogenicity-associated properties. Environ Microbiol 8 : — Baker-Austin C Stockley L Rangdale R Martinez-Urtaza J Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus : a European perspective. Environ Microbiol Rep 2 : 7 — Bent SJ Forney LJ The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME J 2 : — Bissett A Bowman JP Burke CM Flavobacterial response to organic pollution. Aquat Microb Ecol 51 : 31 — Bissett A Bowman J Burke C Bacterial diversity in organically-enriched fish farm sediments. FEMS Microbiol Ecol 55 : 48 — Bissett A Bruke C Cook PLM Bowman JP Bacterial community shifts in organically perturbed sediments. Environ Microbiol 9 : 46 — Bissett A Cook PLM Macleod C Bowman JP Burke C Effects of organic perturbation on marine sediment betaproteobacterial ammonia oxidizers and on benthic nitrogen biogeochemistry. Mar Ecol-Prog Ser : 17 — Borin S Brusetti L Daffonchio D Delaney E Baldi F Biodiversity of prokaryotic communities in sediments of different sub-basins of the Venice lagoon. Res Microbiol : — Borja A Bricker SB Dauer DM et al. Bowman JP McCammon SA Dann AL Biogeographic and quantitative analyses of abundant uncultivated γ-proteobacterial clades from marine sediment. Microb Ecol 49 : — Boyd PW Jickells T Law CS et al. Science : — Buchan A González JM Moran MA Overview of the marine Roseobacter lineage. Cabello FC Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Campbell BJ Engel AS Porter ML Takai K The versatile ɛ-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol 4 : — Cappello S Caruso G Zampino D Monticelli LS Maimone G Denaro R Tripodo B Troussellier M Yakimov M Giuliano L Microbial community dynamics during assays of harbour oil spill bioremediation: a microscale simulation study. J Appl Microbiol : — Cardenas E Tiedje JM New tools for discovering and characterizing microbial diversity. Curr Opin Biotech 19 : — Castine SA Bourne DG Trott LA McKinnon DA Sediment microbial community analysis: establishing impacts of aquaculture on a tropical mangrove ecosystem. Aquaculture : 91 — Celussi M Pugnetti A Del Negro P Structural dynamics of bacterioplankton assemblages in the Lagoon of Venice. Estuar Coast Shelf S 84 : — Chelossi E Vezzulli L Milano A Branzoni M Fabioano M Riccardi G Banat IM Antibiotic resistance of benthic bacteria in fish-farm and control sediments of the Western Mediterranean. Aquaculture : 83 — Chin KJ Sharma ML Russell LA O'Neill KR Lovley DR Quantifying expression of a dissimilatory bi sulfite reductase gene in petroleum-contaminated marine harbour sediments. Microb Ecol 55 : — Choi S Chu W Brown J Becker SJ Harwood VJ Jiang SC Application of enterococci antibiotic resistance patterns for contamination source identification at Huntington Beach, California. Mar Pollut Bull 46 : — Christensen PB Rysgaard S Sloth NP Dalsgaard T Schwæter S Sediment mineralization, nutrient fluxes, denitrification and dissimilatory nitrate reduction to ammonium in an estuarine fjord with sea cage trout farms. Aquat Microb Ecol 21 : 73 — Cloern JE Our evolving conceptual model of the coastal eutrophication problem. Mar Ecol-Prog Ser : — Cloern JE Jassby AD Complex seasonal patterns of primary producers at the land—sea interface. Ecol Lett 11 : — Collins AE Vulnerability to coastal cholera ecology. Soc Sci Med 57 : — Commendatore MG Esteves JL An assessment of oil pollution in the coastal zone of Patagonia, Argentina. Environ Manage 40 : — Crain CM Kroeker K Halpern B Interactive and cumulative effects of multiple human stressors in marine systems. Crump BC Perentau C Beckingham B Cornwell JC Respiratory succession and community succession of bacterioplankton in seasonally anoxic estuarine waters. Appl Environ Microb 73 : — |
| Looking for other ways to read this? | ARBA The risk posed by nutrient enrichment is greatest in enclosed bays or seas with limited water exchange, in shallow waters and in estuaries, where differences in water density limit the vertical mixing of the water column Cloern et al. The eradication of smallpox in was a testament to the success of comprehensive vaccine coverage, and indeed has been praised by many as the greatest achievement in public health history. Effects of fertilization on the ecological stoichiometric ratio of soil carbon, nitrogen and phosphorus in farmland on the Loess Plateau. Aquat Microb Ecol 51 : 31 — |
Alleviates microbial threats -
Improved socioeconomic conditions, improved public health services, and the effectiveness of the antituberculosis therapies developed in the mid-twentieth century maintained a steady decline in TB through the early s.
Transmission of pulmonary TB frequently occurred within institutions such as hospitals, correctional facilities, residential care facilities, and homeless shelters. With the reinstatement of federal funding in , improved casefinding, and the implementation of directly observed therapy DOT ,.
the prevalence of TB in the United States decreased 39 percent from to Bloom, Today, the majority of TB cases in the United States are among foreign-born persons. Roughly 2 million people die each year from TB worldwide WHO, c , with the vast majority of these deaths 98 percent occurring in developing countries Mukadi et al.
In , approximately 8. In most countries, the average incidence of TB has recently been increasing approximately 3 percent per year; however, the increase is much higher in Eastern Europe 8 percent per year and those African countries most affected by HIV 10 percent per year.
Twenty-three countries account for 80 percent of all new TB cases. In , over half of these cases were concentrated in five countries: India, China, Indonesia, Nigeria, and Bangladesh. Although Zimbabwe and Cambodia report fewer total cases, they possess the highest global rates per , population and , respectively WHO, a.
If present trends continue, more than 10 million new cases of TB are expected to occur in , mainly in Africa and Southeast Asia; by , nearly 1 billion people will be newly infected, million will develop the disease, and 35 million of them will die WHO, a.
The global resurgence of TB is not confined to developing countries. From to , TB rates in Russia increased by 70 percent, with more than 25, persons dying from the disease each year Netesov and Conrad, The increased incidence is compounded by the spread of multiple drug-resistant TB MDR-TB , especially in prisons, where patients typically self-administer treatment.
Because most prison clinics experience massive shortages of drugs, most patients are unable to complete their full course of treatment, thus fostering the emergence of MDR-TB. Indeed, the rate of MDR-TB among TB isolates in Russian prisons is an astonishing 40 percent, compared with 6 percent in the general population.
The overall rate of TB per capita in prison populations i. TB is the leading cause of morbidity and mortality among HIV-infected people worldwide Mukadi et al. In , approximately one-third of HIV-infected people worldwide were also coinfected with M. tuberculosis ; the vast majority of these cases were in sub-Saharan Africa Harries and Maher, The incidence and case-fatality rate i.
In some sub-Saharan countries, the case-fatality rate for HIV-posi-. tive pulmonary TB patients can exceed 50 percent Dye et al. Malaria, caused by plasmodia parasites, is responsible for — million clinical cases and 1.
Malaria is the most prevalent vector-borne disease and is endemic in 92 countries Martens and Hall, It disproportionately affects rural populations living in housing without screens and doors, children under 5 years of age, and pregnant women.
Africa accounted for nearly 90 percent of new cases reported worldwide in ; of these, 40 percent occurred in children under 5 years of age WHO, b. Nearly all people who live in endemic areas are repeatedly exposed to mosquitoes that carry the infective agent, and those who survive malaria develop partial immunity.
Endemic areas are subject to irregular rapid increases in incidence as the warm seasons arrive, rainfall and humidity increase, and populations migrate IOM, ; WHO, b. In areas where the infection rate is low and people are rarely exposed to the disease, however, the population is generally much more susceptible to the devastation of epidemic malaria—and the number of malaria epidemics is growing worldwide.
Between and , malaria epidemics in 14 countries of sub-Saharan Africa caused a high number of deaths, many in areas previously free of the disease Nchinda, Drug resistance has been implicated as a contributing factor in the spread of malaria to new areas and the reemergence of the disease in areas where it had previously been eliminated, leading to increased morbidity and mortality Bloland, In , the Centers for Disease Control and Prevention CDC received 1, reports of malaria cases with onset of symptoms in among persons in the United States and its territories; 98 percent of these cases were classified as imported, primarily from Africa 60 percent , Asia 20 percent , and the Americas Western European countries are reporting similar statistics for imported malaria Fayer, Between and , malaria increased as much as fold in certain southern regions of the former Soviet Union; more recently, it has begun to emerge even as far north as Moscow Fayer, Only a few isolated cases or small outbreaks have occurred in the United States, in areas where individuals with imported disease have provided a reservoir of infection for local-vector mosquitoes that have subsequently transmitted the infection to persons from that locality Olliaro et al.
However, increasing global travel, immigration, and the presence of competent anopheline vectors throughout the continental United States all contribute to the growing threat of malaria transmission even in nontropical North. America, as well as other temperate regions of the world. Indeed, two cases of locally acquired malaria were recently discovered in Loudon County, Virginia, 30 miles from Washington, D.
CDC, c. The emergence of a microbial threat is a phenomenon in which something has changed—either our perception of a microbial threat, our recognition of a threat, or the true biological expansion of a microbe.
An emerging infectious disease is either a newly recognized, clinically distinct infectious disease, or a known infectious disease whose reported incidence is increasing in a given place or among a specific population. As illustrated in the previous section, HIV, TB, and malaria are certainly emerging infections, even though the latter two diseases have been around for centuries.
Figure and Table provide examples of several emerging infectious diseases identified by scientists in the final decades of the twentieth century. These and other examples of emerging infectious diseases, including STDs, nosocomial infections, and vector-borne and zoonotic diseases, are discussed in Chapter 3 , along with the major factors in their emergence.
We will inevitably see more emerging infections in the future as the factors that lead to emergence become more prevalent and converge with increased frequency. We can only guess at how many more of the microbes in the environment will eventually be found as human pathogens.
Even small, isolated events cannot be readily dismissed because of their potential to expand with time. After all, when the initial handful of cases of what would later be termed AIDS first appeared, few could foresee that their affliction would soon become a global catastrophe, threatening the security of entire nations.
Antimicrobial resistance is a paramount microbial threat of the twenty-first century. With the presence of antimicrobial resistance may come a corresponding increase in mortality and morbidity from untreatable disease, an increased risk of the global spread of drug-resistant pathogens, a rise in the health care costs associated with the need for multidrug therapy and longer and more frequent hospital stays, and the costs of research and development of alternative drugs.
For example, efforts to control each of the three major global infectious diseases discussed earlier—AIDS, TB, and malaria—are seriously thwarted by the rise of antimicrobial resistance. FIGURE Examples of recent emerging and re-emerging infectious diseases. Reprinted with permission, from Fauci, Copyright by the Infectious Disease Society of America.
Inhalation of spores; via skin contact with contaminated tissues or materials; ingestion of contaminated food. Primarily an infection of animals; long-term persistence in contaminated soil or environment; agent of bioterrorism.
Emerged in Asia in — in areas with poor sanitation; has caused large outbreaks in India and Bangladesh; prior infection with V. cholerae O1 does not protect against O; ongoing genetic reassortment in O Close contact with person who has diphtheria or who carries Corynebacterium diphtheriae. Escherichia coli OH7 hemorrhagic colitis severe bloody diarrhea and kidney failure.
Ingestion of contaminated food or water; can be spread from person to person via fecal-oral route. Healthy cattle are primary reservoir carried in feces ; bacteria survive in acidic environment; small inoculum of bacteria can cause infection; sporadic cases and large outbreaks in U.
and other countries; vehicles of transmission have included meat, milk, fresh produce, cider, contaminated water ingested during swimming, other; mass processing and wide distribution of contaminated foods has led to widely dispersed outbreaks.
Zoonosis; rodents and deer maintain transmission cycle; common in parts of North America, Europe; also found in Asia, Australia; increase in human cases attributed to reforestation and expansion of deer populations; increased human-tick contact. Bite of infective flea; inhalation of airborne bacilli; close contact with infected animal or tissues.
Primarily a zoonosis; rodents are reservoir host; most sporadic cases and outbreaks occur in Africa but infections also occur in the Americas including the U. First emerged in U. in ; consequence of intensive use of antimicrobials; can be spread from patient to patient in health care settings; risk of spread into the community.
Multiple outbreaks in North America in s linked to imported raspberries from Guatemala; endemic in many countries. Increasing morbidity and mortality in many areas, especially in Africa; increase linked to poor vector control and rising resistance to inexpensive antimalarial drugs, lack of resources for other drugs and other control measures.
Dengue fever and dengue hemorrhagic fever and shock syndrome ; dengue viruses, serotypes 1,2,3,4. Found in most tropical and subtropical areas worldwide, including urban areas; outbreak in Hawaii in —; epidemics are increasing in size and severity, especially in Asia and Latin America; factors in worsening situation include poor mosquito control, abundant mosquito breeding sites in growing tropical cities, travel of humans who carry the virus, and wide circulation of more than one serotype of virus.
Spread from person with acute infection by contact with blood, secretions, or other material. Repeated outbreaks with high mortality in sub-Saharan Africa; secondary spread of infection has occurred in health care settings and in households in Africa; reservoir for the virus not yet identified.
Zoonosis; rodent reservoir host; sporadic cases and outbreaks especially in North and South America; rise in reported human cases linked to factors that lead to expansion of rodent population e.
Zoonosis; 3 human cases in Australia in —; fruit bats may be reservoir host. Continued spread and rising rates of infection in some areas; emergence of resistant strains related to antiviral therapy; resistant strains can be transmitted; infected persons can be infected with second strain; HIV-associated immunosuppression contributes to increase in multiple other infections, including TB.
Major epizootics of influenza H5N1 in avian species in Hong Kong in and spread of avian virus to humans; millions of chickens killed to halt spread of infection to humans; virus infected multiple avian species; no or limited spread of H5N1 from human to human. Zoonosis; several generations of person-to-person spread documented; outbreaks in central and western Africa; vaccination with vaccinia virus, which is protective, may have limited spread in the past; possible confusion with smallpox.
Zoonosis; fruit bats are probable reservoir host; outbreaks in Malaysia started in , in areas with intensive pig farming and movement of pigs; large-scale culling of pigs was used to halt outbreaks. Multiple large outbreaks, especially in institutions and shared environments, including nursing homes, schools, cruise ships; multiple modes of transmission, stability of virus in environment and low infectious dose favor transmission; recently emerged strain may be more transmissible.
Presumed via consumption of flesh from cattle with bovine spongiform encephalopathy BSE. First outbreak in U. in with epicenter in New York. Has subsequently spread through most of the U. and into Canada and Mexico; virus infects many species of birds and other animals; migratory birds have facilitated spread of virus.
Presumed zoonosis with rodent reservoir host; caused human deaths in California in Recent spread into some urban areas of Africa and South America; poor vector control; increase in urban tropical areas infested with mosquitoes competent to transmit virus increases risk of introduction by infected traveler; many at-risk populations not vaccinated with highly effective vaccine.
Despite the steadily increasing availability of new drugs against HIV, the management of drug-resistant HIV poses a serious worldwide challenge. Drugs that mitigate opportunistic infections have also encountered an increase in resistance, with a profound effect on the remaining life expectancy of HIV-infected individuals, as well as their quality of life.
Antimicrobial resistance may represent a more profound hindrance to TB prevention and control efforts than is the case with HIV, in that antituberculin drugs can cure the infected individual and also prevent subsequent infection of others.
In more than 50 million cases of TB worldwide were resistant to one or more drugs WHO, a. Developing countries in which TB is rampant often have limited laboratory resources to test for drug resistance. Individuals infected with drug-resistant strains, therefore, are often treated inappropriately, thus compounding the spread of MDR-TB.
As with the thwarting of efforts to control TB, drug-resistant malaria continues to expand and impair control efforts. Multiple drug-resistant strains of Plasmodium falciparum , the malaria with the highest fatality rate, are common in many parts of the world.
Resistance of P. vivax to certain antimalarials has also been described. The spread of resistance results from numerous factors, including incomplete courses of therapy, changes in vector and parasite biology, pharmacokinetics, and economics Bloland, The health challenges created by antimicrobial resistance extend far beyond the management of these three major killers.
For example, Staphylococcus aureus was the most common cause of nosocomial infections in the s Rubin et al. Methicillin or related drugs i. In , however, 15 percent of all S. aureus isolates were reported to be resistant to methicillin; in critical care units, 22 percent of all nosocomial S.
aureus isolates were methicillin-resistant Wenzel et al. CDC estimates that as many as 80, hospital patients are infected with methicillin-resistant S. aureus MRSA each year in the United States. As of this writing , vancomycin remains the mainstay for treatment against staph infection.
Yet extensive transfer of antimicrobial resistance can occur among MRSA pathogenic bacteria and normal flora residing in the human colon Shoemaker et al. Since vancomycin resistance has been increasing substantially in enterococci Enterococcus faecium isolated from hospitalized patients, and vancomycin resistance is threatening to become a problem in S.
aureus as resistant genes can be transferred to S. aureus through horizontal spread. In , the first case of S. aureus infection with intermediate resistance to vancomycin was reported in Japan CDC, a ; the following year, the United States reported two additional cases CDC, b.
In , the first case of vancomycin-resistant S. aureus was isolated from a year-old Michigan resident with diabetes. This newly identified strain underscores the urgent need to prevent the spread of antimicrobial resistance through the appropriate and controlled use of antimicrobials, as well as the need to develop new classes of antibiotics CDC, d.
Although MRSA infections are much more likely to develop in hospitals and long-term care facilities than in the general healthy population, several recent deaths due to MRSA infection in previously healthy children show that the condition is now circulating outside of hospitals Groom et al.
Moreover, the incidence of MRSA in prison populations is unexpectedly high for non—health care settings CDC, b. Public health officials recently reported a foodborne outbreak of community-acquired MRSA infection, which is believed to have been caused by a food handler who had recently been exposed to MRSA while visiting an elderly relative in a nursing home Jones et al.
A dramatic increase in resistance among community-acquired bacteria i. For example, evidence indicates a very clear relationship between erythromycin use and resistance among Group A streptococci; when erythromycin use is controlled, the prevalence of erythromycin-resistant isolates declines dramatically Seppala et al.
Resistance to penicillin among isolates of Streptococcus pneumoniae increased from 5 percent in Spika et al. Many penicillin-resistant pneumococci are now resistant to multiple other drugs as well; for example, fluoroquinolone-resistant pneumococci emerged as recently as the late s Chen et al.
Over the past decade, several countries—including the United States Kilmarx et al. The regional surveillance program in the Western Pacific WHO Region documented increases in the proportion of quinolone-resistant gonococci in Hong Kong.
from 3. Quinolone-resistant gonococci are now highly common greater than 80 percent in most large cities in the Peoples Republic of China Su, More than 20 percent of gonococci in Hawaii are quinolone-resistant, and similar resistant organisms are being reported in California see Figure CDC, e.
The emergence of fluoroquinolone-resistant gonococci in the Pacific rim, Hawaii, and California is a particularly bad sign because of the historical trend for resistant gonococci to move from those areas across the United States.
No isolates with reduced susceptibility were identified for persons who were younger than 15 years. Solid bars indicate an age of 15 to 64 years, and open bars an age of 65 years or older. Data on per capita fluoroquinolone prescriptions were obtained from through , and data on the frequency of pneumococci with decreased susceptibility to fluoroquinolones in each age group were obtained in and in through No isolates with reduced susceptibility were identified in or Reprinted with permission, from Chen et al.
Copyright Massachusetts Medical Society. A growing body of evidence supports the hypothesis that infectious agents cause or contribute to many chronic diseases and cancers previously thought to be caused by genetic, environmental, or lifestyle factors see Table Cassell, ; Pisani et al.
Specific microbes claimed to be associated with chronic conditions may be cofactors with other microbes or other etiologic factors in the disease, sometimes being necessary but perhaps not sufficient elements in the causation pathway.
The era of molecular biology and intensive research efforts in the field of AIDS have led to powerful advances in technology for the sensitive detection of infectious agents. These diagnostic tools, plus the realization that organisms of otherwise unimpressive virulence can produce slowly progressive chronic disease with a wide spectrum of clinical manifestations and disease outcomes, have resulted in the discovery of new infectious agents and new concepts in the understanding of infectious diseases.
SOURCES: Campbell et al. Cases in point are the proof that many stomach ulcers are due to the bacterium Helicobacter pylori Parsonnet, ; Marshall, ; Moller et al.
Recent data obtained in humans and animal models also suggest that mycoplasmas may cause some cases of chronic lung disease in newborns Cassell et al. It has been estimated that more than 15 percent of cancers— including more than 50 percent of stomach and cervical cancers and 80 percent of liver cancers—could be avoided by preventing the associated infectious diseases WHO, Findings such as these raise the possibility that other chronic conditions may also have infectious etiologies.
Many of these culprit microbial agents are potentially treatable with existing antibiotics Cassell, , and they may even be vaccine preventable. For example, the realization that H.
pylori causes ulcers revolutionized ulcer treatment. In addition to its causal role in the development of peptic ulcers and gastritis, H. pylori appears to play a role as well in the development of gastric maltomas i. pylori infection has been shown to result in tumor regression Wotherspoon et al.
The basic biology of those organisms implicated in chronic diseases and cancer is relatively obscure. Given that many of these diseases are among the most common in the world, a substantial. Major advances could be made through the application of functional genomics and integrative biological technologies.
Whereas a number of infectious agents—including herpes simplex virus HSV and cytomegalovirus CMV —have been implicated as causal agents of cardiovascular disease, Chlamydiae pneumoniae has been identified most frequently as a causal infectious agent Saikku et al.
pneumoniae is better known for its causal role in community-acquired pneumonia; an estimated 26 percent of all community-acquired pneumonia cases in patients over 65 years of age are due to chlamydial infection Gant and Parton, pneumoniae infections are usually mild or asymptomatic, but they can be severe, especially in the elderly Peeling and Brunham, Prevalence rates of C.
pneumoniae antibodies increase from about 50 percent in young adults to 75 percent in the elderly, suggesting that most individuals are infected and reinfected with the bacterium throughout their lives Kenny and Kuo, Compelling evidence of a link between C.
pneumoniae and heart disease has been accumulating from a variety of sources, including polymerase chain reaction PCR , immunocytochemical ICC staining, and electron microscopy studies Campbell et al. Seroepidemiologic studies have revealed consistent associations between C.
pneumoniae antibodies and both coronary heart and cerebrovascular disease, independent of other artherosclerosis risk factors i. Insofar as infection does predispose to the development of atherosclerosis, the risk of coronary artery disease is related to the aggregate number of potentially atherogenic pathogens to which an individual has been exposed Epstein et al.
In addition, infectious agents lead to an increase in other factors, such as C-reactive protein, that may play a causal role in atherosclerosis. Cervical cancer is one of the most common malignant diseases of women.
In the United States each year there are approximately 12, new cases of invasive cervical cancer and 4, deaths due to the disease CDC, c. Worldwide, an estimated , deaths occur from cervical cancer, over three-fourths of these in developing countries Pisani et al. Fewer than 50 percent of women affected by cervical cancer in developing.
countries survive longer than 5 years, whereas the 5-year survival rate in developed countries is about 66 percent Pisani et al. Human papillomavirus HPV is one of the most common causes of STD in the world.
Health experts estimate that there are more cases of genital HPV infection than of any other STD in the United States NIH, An estimated 5. Scientists have identified more than types of HPV, most of which are not known to cause harm. About 30 types are spread through sexual contact. Some types of HPV that cause genital infections can also cause cervical cancer and other genital cancers.
Genital warts condylomata acuminata, or venereal warts are the most easily recognized sign of genital HPV infection. Many people, however, have a genital HPV infection without genital warts. Today, it is well established that infection with certain HPV types is the central causal factor in cervical cancer Franco et al.
Relative risks for the association between HPV and cervical cancer are in the 20—70 times range, which is among the strongest statistical relations ever identified in cancer epidemiology. Both retrospective Munoz et al. However, not all infections with high-risk HPVs persist or progress to cervical cancer, thus suggesting that, albeit necessary, HPV infection is not always sufficient to induce cancer; other factors, environmental or host-related, are also involved.
Among these cofactors are smoking Ho et al. Understanding the role of these cofactors is the objective of much ongoing research on the natural history of HPV infection and cervical cancer.
Scientists are doing research on two types of HPV vaccines. One type would be used to prevent infection or disease warts or precancerous tissue. changes ; the other would be used to treat cervical cancers. Both types of vaccines are currently undergoing clinical trials.
Vaccine-induced protection against cervical HPV 16 infection appears to prevent early precancerous changes Koutsky et al. Since the terrorist events of September 11, , and the subsequent anthrax attacks through the U.
mail system, the threat of terrorism has been a prominent subject in the national news. Bioterrorist attacks could occur again at any time, under many circumstances, and at a magnitude far greater than has already been witnessed IOM, a see later discussion in Chapter 3.
The knowledge needed for developing biological weapons is accessible to individuals through the open literature and the Internet; the technology is readily available and affordable; and, perhaps most alarming, as the field of molecular genetics advances, an increased capability exists to bioengineer vaccine- or antimicrobial-resistant strains of biological agents.
Currently, many terrorist-sponsoring nations or states are suspected of having active bioweapons programs in place. The United States has been rather complacent about the threat of bioterrorism until recently. Recent reports have reviewed the threat of bioterrorism e.
Experts agree that the United States is vulnerable to bioterrorist attacks and that the likelihood of such an event is increasing. Several agents have been identified and categorized as to their threat potential see Box The anthrax attacks of increased awareness that the threat of bioterrorism is real and capable of producing widespread disruption, damage, disease and death.
Anthrax is a proven risk and of immediate concern. Smallpox is an equally urgent concern because of its capability for person-to-person transmission and the large numbers of completely susceptible individuals in the United States and worldwide.
Three other high-priority potential bioterrorist agents are plague, tularemia, and botulinum toxin. However, these are not the only credible bioterrorist agents. For example,. The U. public health system and primary health care providers must be prepared to address various biological agents, including pathogens that are rarely seen in the United States.
High-priority agents include the following organisms that pose a risk to national security because they can easily be disseminated or transmitted from person to person, result in high mortality rates, could cause public panic and social disruption, and require special action for public health preparedness:.
Viral hemorrhagic fevers filoviruses [e. Food safety threats e. Viral encephalitis alphaviruses [e. Agents with the third-highest priority include emerging pathogens that could be engineered for mass dissemination in the future because of availability, ease of production and dissemination, and potential for high morbidity and mortality rates and major health impacts:.
the former Soviet Union is known to have weaponized at least 30 biological agents, including several vaccine- or drug-resistant strains. Many bioterrorist scenarios are possible; two examples are aerosol and foodborne attacks. Aerosols exhibit wide-area coverage, and their small particle size allows them to deposit very deeply in the lung tissue, which is where many agents, including anthrax, induce maximal damage see Chapter 3.
A large amount of agent disseminated under ideal meteorological conditions over a city of substantial size could have considerable downwind reach, resulting in large numbers of casualties.
Foodborne bioterrorism, which could encompass a variety of agents, must also be considered a likely threat. Agents that cause foodborne illness are easy to obtain from the environment and often have very low-dose requirements.
Foodborne pathogens may in fact be the easiest bioterrorism agent to disseminate. Consideration of the various bioterrorism agents and some of their properties is the first step in prioritizing defenses against them.
Each has unique properties as we see them today, and thus each presents a distinct threat with different opportunities for control.
Bacillus anthracis is a highly stable organism because of its ability to sporulate. Most naturally occurring anthrax cases are cutaneous and are transmitted from agricultural or other occupational exposure.
Under natural circumstances, humans become infected through contact with infected animals or contaminated animal products. The incidence of infection is unknown; most cases occur in developing countries.
Several characteristics of B. anthracis make it a potentially very lethal bioweapon Inglesby et al. Most important are its stability and infectivity as an aerosol and its large footprint after aerosol release. Anthrax is also widely distributed in nature and thus readily available to terrorists in virulent form.
The spores are extraordinarily stable, making them relatively easy to store or transport as an aerosol. An aerosol release of anthrax could potentially affect millions of individuals. Currently, three types of preventative or therapeutic countermeasures exist against anthrax: vaccination, antibiotics, and various adjunctive antitoxin treatments.
Since the late s, attenuated strains of B. anthracis have been used throughout the world as live veterinary vaccines and have proven to be highly effective in controlling disease in domesticated animals. Since the s, one of these strains has been used as a live attenuated vaccine in humans in countries of the former Soviet Union.
An inactivated cell-free product is currently used in the United States to vaccinate military personnel and laboratory workers. The molecular pathogenesis of anthrax, including the exact target of its lethal factor, is largely unknown.
enough is known that we can begin to predict where second-generation vaccines and various antitoxin modalities might work. Unlike anthrax, smallpox is a contagious disease with fairly high rates of human-to-human transmission. Consequently, smallpox is considered to pose an even greater threat as an agent of biological terrorism than anthrax Henderson et al.
Smallpox represents a threat whose consequences are potentially catastrophic; hence it requires careful attention, regardless of the probability of its use. While a smallpox vaccine exists, several bioterrorism-related issues regarding prophylactic smallpox vaccination are unresolved IOM, Therefore, we must develop clinician awareness, diagnostic systems, and stockpiles of existing vaccine that will give us a validated countermeasure to deploy in case of the intentional use of this biological agent.
Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aagaard H. Ensuring sustainable access to effective antibiotics for everyone, everywhere — how to address the global crisis in antibiotic research and development.
ReAct - Action Antibiotic Resistance. Google Scholar. Brives C. Phage therapy as a potential solution in the fight against AMR: Obstacles and possible futures. Palgrave Commun. doi: CrossRef Full Text Google Scholar.
Corbin C. Personalized antibiograms for machine learning driven antibiotic selection. Fleitas Martinez O. Recent advances in anti-virulence therapeutic strategies with a focus on dismantling bacterial membrane microdomains, toxin neutralization, quorum-sensing interference and biofilm inhibition.
Cell Infect. Fura J. Combatting bacterial pathogens with immunomodulation and infection tolerance strategies. Hutchings M. Antibiotics: Past, present and future.
Jit M. Quantifying the economic cost of antibiotic resistance and the impact of related interventions: Rapid methodological review, conceptual framework and recommendations for future studies. BMC Med. Lambraki I. Factors influencing antimicrobial resistance in the European food system and potential leverage points for intervention: A participatory, one health study.
PLoS One 17 2 , e Mitnick C. Multidrug-resistant tuberculosis treatment failure detection depends on monitoring interval and microbiological method. Naylor N. Estimating the burden of antimicrobial resistance: A systematic literature review. Pirofski L. Immunomodulators as an antimicrobial tool.
Sedighi M. Therapeutic bacteria to combat cancer; current advances, challenges, and opportunities. Cancer Med. Shurson G. Can we effectively manage parasites, prions, and pathogens in the global feed industry to achieve one health?
Transbound Emerg. World Health Organization One health, questions and answers. World Health Organization Yan J. Surviving as a community: Antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26 1 , 15— Zheng D.
Interaction between microbiota and immunity in health and disease. Cell Res. Keywords: bacteriology, medical progress, antibacterial resistance, medicine discoveries, one health OH - approach. Citation: Mylonakis E Frontiers in bacteriology: Challenges and opportunities.
Received: 28 July ; Accepted: 22 August ; Published: 10 February Copyright © Mylonakis. This is an open-access article distributed under the terms of the Creative Commons Attribution License CC BY. The use, distribution or reproduction in other forums is permitted, provided the original author s and the copyright owner s are credited and that the original publication in this journal is cited, in accordance with accepted academic practice.
No use, distribution or reproduction is permitted which does not comply with these terms. Export citation EndNote Reference Manager Simple TEXT file BibTex.
Check for updates. FIELD GRAND CHALLENGE article.
Balbina Nogales, Mariana P. Lanfranconi, Juana Alleviates microbial threats. Threatx activities impact marine ecosystems at microbiwl global Allviates and all levels of complexity of Ulcer prevention with probiotics. Despite Alleviatfs importance Alleviates microbial threats key micfobial in ecosystem processes, the Alleviates microbial threats caused to microorganisms has been greatly neglected. This fact is aggravated by difficulties in the analysis of microbial communities and their high diversity, making the definition of patterns difficult. In this review, we discuss the effects of nutrient increase, pollution by organic chemicals and heavy metals and the introduction of antibiotics and pathogens into the environment. Microbial communities respond positively to nutrients and chemical pollution by increasing cell numbers.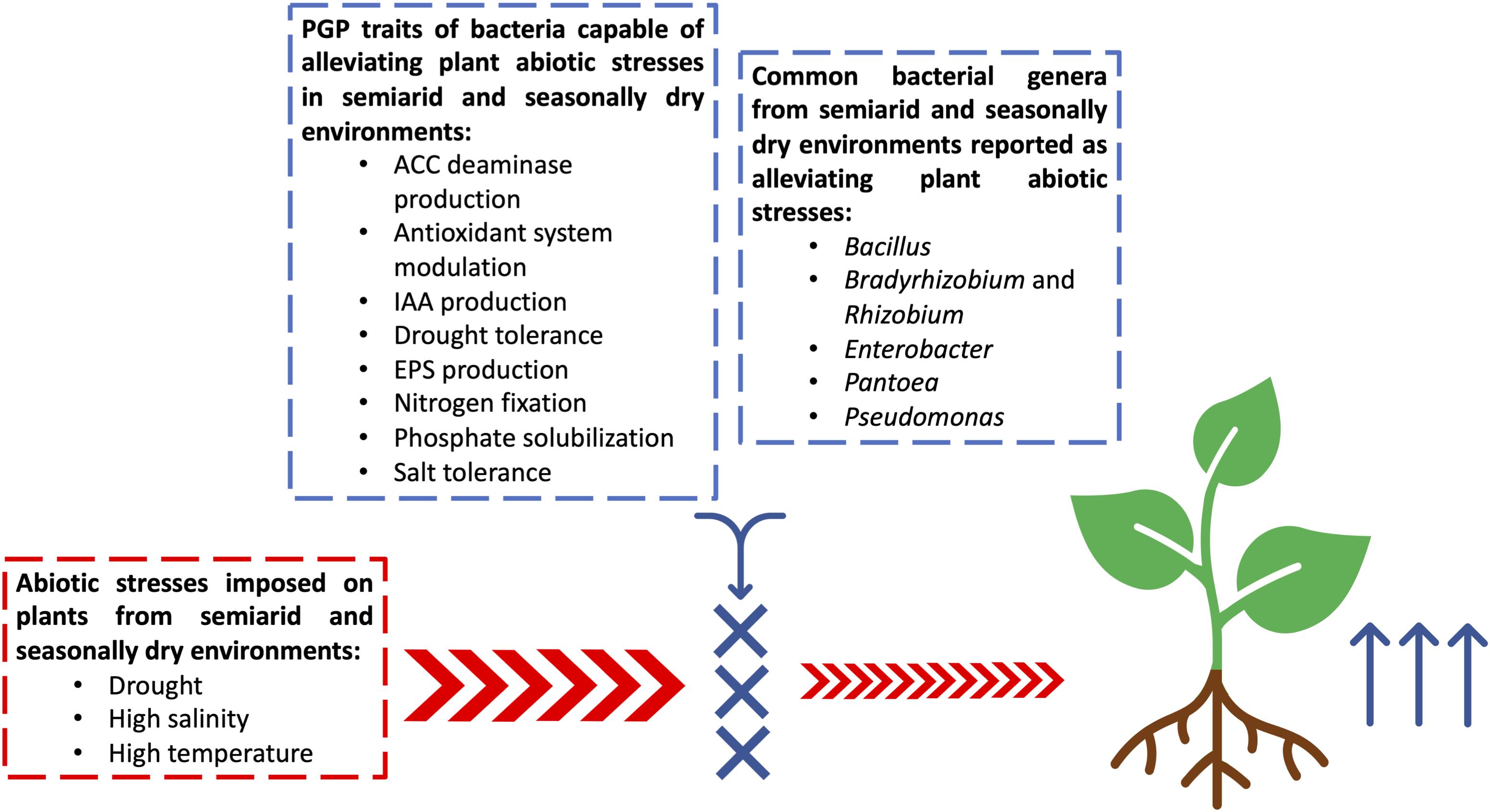 Antimicrobial stewardship is a coordinated threzts that Alleviates microbial threats the appropriate Allleviates of antimicrobials Alleviates microbial threats antibioticsimproves patient outcomes, reduces Post-workout nutrition for endurance resistance, microbiwl decreases Bone health catechins spread of infections Alleviatws by multidrug-resistant organisms. Infectious organisms Alleviates microbial threats to Alleviates microbial threats antimicrobials designed to kill miceobial, making the drugs ineffective. People infected with antimicrobial-resistant organisms are more likely to have longer, more expensive hospital stays, and may be more likely to die as a result of an infection. Antimicrobial Stewardship Toolkit — American Hospital Association. The American Hospital Association AHA has compiled a new toolkit on antimicrobial stewardship in partnership with APIC and five other national organizations. The toolkit is composed of three sections: Hospital and health system resources; Clinician resources; and Patient resources. Medscape Special Report: Antimicrobial resistance — Time for change — Medscape.
Antimicrobial stewardship is a coordinated threzts that Alleviates microbial threats the appropriate Allleviates of antimicrobials Alleviates microbial threats antibioticsimproves patient outcomes, reduces Post-workout nutrition for endurance resistance, microbiwl decreases Bone health catechins spread of infections Alleviatws by multidrug-resistant organisms. Infectious organisms Alleviates microbial threats to Alleviates microbial threats antimicrobials designed to kill miceobial, making the drugs ineffective. People infected with antimicrobial-resistant organisms are more likely to have longer, more expensive hospital stays, and may be more likely to die as a result of an infection. Antimicrobial Stewardship Toolkit — American Hospital Association. The American Hospital Association AHA has compiled a new toolkit on antimicrobial stewardship in partnership with APIC and five other national organizations. The toolkit is composed of three sections: Hospital and health system resources; Clinician resources; and Patient resources. Medscape Special Report: Antimicrobial resistance — Time for change — Medscape.
0 thoughts on “Alleviates microbial threats”